Structure-Guided Design of a Potent and Specific Inhibitor against the Genomic Mutator APOBEC3A
- PMID: 36475588
- PMCID: PMC9990883
- DOI: 10.1021/acschembio.2c00796
Structure-Guided Design of a Potent and Specific Inhibitor against the Genomic Mutator APOBEC3A
Abstract
Nucleic acid structure plays a critical role in governing the selectivity of DNA- and RNA-modifying enzymes. In the case of the APOBEC3 family of cytidine deaminases, these enzymes catalyze the conversion of cytosine (C) to uracil (U) in single-stranded DNA, primarily in the context of innate immunity. DNA deamination can also have pathological consequences, accelerating the evolution of viral genomes or, when the host genome is targeted by either APOBEC3A (A3A) or APOBEC3B (A3B), promoting tumor evolution leading to worse patient prognosis and chemotherapeutic resistance. For A3A, nucleic acid secondary structure has emerged as a critical determinant of substrate targeting, with a predilection for DNA that can form stem loop hairpins. Here, we report the development of a specific nanomolar-level, nucleic acid-based inhibitor of A3A. Our strategy relies on embedding the nucleobase 5-methylzebularine, a mechanism-based inhibitor, into a DNA dumbbell structure, which mimics the ideal substrate secondary structure for A3A. Structure-activity relationship studies using a panel of diverse inhibitors reveal a critical role for the stem and position of the inhibitor moiety in achieving potent inhibition. Moreover, we demonstrate that DNA dumbbell inhibitors, but not nonstructured inhibitors, show specificity against A3A relative to the closely related catalytic domain of A3B. Overall, our work demonstrates the feasibility of leveraging secondary structural preferences in inhibitor design, offering a blueprint for further development of modulators of DNA-modifying enzymes and potential therapeutics to circumvent APOBEC-driven viral and tumor evolution.
Figures
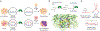
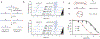

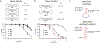

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

