The IRE1α-XBP1 Signaling Axis Promotes Glycolytic Reprogramming in Response to Inflammatory Stimuli
- PMID: 36475773
- PMCID: PMC9973330
- DOI: 10.1128/mbio.03068-22
The IRE1α-XBP1 Signaling Axis Promotes Glycolytic Reprogramming in Response to Inflammatory Stimuli
Abstract
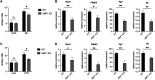
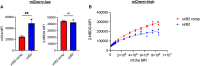
Immune cells must be able to adjust their metabolic programs to effectively carry out their effector functions. Here, we show that the endoplasmic reticulum (ER) stress sensor Inositol-requiring enzyme 1 alpha (IRE1α) and its downstream transcription factor X box binding protein 1 (XBP1) enhance the upregulation of glycolysis in classically activated macrophages (CAMs). The IRE1α-XBP1 signaling axis supports this glycolytic switch in macrophages when activated by lipopolysaccharide (LPS) stimulation or infection with the intracellular bacterial pathogen Brucella abortus. Importantly, these different inflammatory stimuli have distinct mechanisms of IRE1α activation; while Toll-like receptor 4 (TLR4) supports glycolysis under both conditions, TLR4 is required for activation of IRE1α in response to LPS treatment but not B. abortus infection. Though IRE1α and XBP1 are necessary for maximal induction of glycolysis in CAMs, activation of this pathway is not sufficient to increase the glycolytic rate of macrophages, indicating that the cellular context in which this pathway is activated ultimately dictates the cell's metabolic response and that IRE1α activation may be a way to fine-tune metabolic reprogramming. IMPORTANCE The immune system must be able to tailor its response to different types of pathogens in order to eliminate them and protect the host. When confronted with bacterial pathogens, macrophages, frontline defenders in the immune system, switch to a glycolysis-driven metabolism to carry out their antibacterial functions. Here, we show that IRE1α, a sensor of ER stress, and its downstream transcription factor XBP1 support glycolysis in macrophages during infection with Brucella abortus or challenge with Salmonella LPS. Interestingly, these stimuli activate IRE1α by independent mechanisms. While the IRE1α-XBP1 signaling axis promotes the glycolytic switch, activation of this pathway is not sufficient to increase glycolysis in macrophages. This study furthers our understanding of the pathways that drive macrophage immunometabolism and highlights a new role for IRE1α and XBP1 in innate immunity.
Keywords: Brucella; endoplasmic reticulum; immunometabolism; innate immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Baumler AJ, Santos RL, Tsolis RM. 2013. PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14:159–170. doi:10.1016/j.chom.2013.07.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
