Multifaceted Interplay between Hfq and the Small RNA GssA in Pseudomonas aeruginosa
- PMID: 36475775
- PMCID: PMC9973299
- DOI: 10.1128/mbio.02418-22
Multifaceted Interplay between Hfq and the Small RNA GssA in Pseudomonas aeruginosa
Abstract
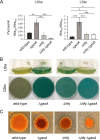
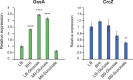
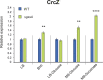
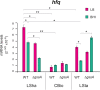
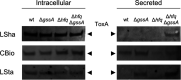
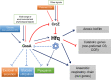
Behind the pathogenic lifestyle of Pseudomonas aeruginosa exists a complex regulatory network of intertwined switches at both the transcriptional and posttranscriptional levels. Major players that mediate translation regulation of several genes involved in host-P. aeruginosa interaction are small RNAs (sRNAs) and the Hfq protein. The canonical role of Hfq in sRNA-driven regulation is to act as a matchmaker between sRNAs and target mRNAs. Besides, the sRNA CrcZ is known to sequester Hfq and abrogate its function of translation repression of target mRNAs. In this study, we describe the novel sRNA GssA in the strain PA14 and its multifaceted interplay with Hfq. We show that GssA is multiresponsive to environmental and physiological signals and acts as an apical repressor of key bacterial functions in the human host such as the production of pyocyanin, utilization of glucose, and secretion of exotoxin A. We suggest that the main role of Hfq is not to directly assist GssA in its regulatory role but to repress GssA expression. In the case of pyocyanin production, we suggest that Hfq interplays with GssA also by converging a positive effect on this pathway. Furthermore, our results indicate that both Hfq and GssA play a positive role in anaerobic growth, possibly by regulating the respiratory chain. On the other hand, we show that GssA can modulate not only Hfq expression at both transcriptional and posttranscriptional levels but also that of CrcZ, thus potentially influencing the pleiotropic role of Hfq. IMPORTANCE The pathogenic lifestyle of the bacterium Pseudomonas aeruginosa, a leading cause of life-threatening infections in the airways of cystic fibrosis patients, is based on the fine regulation of virulence-associated factors. Regulatory small RNAs (sRNAs) and the RNA-binding protein Hfq are recognized key components within the P. aeruginosa regulatory networks involved in host-pathogen interaction. In this study, we characterized in the highly virulent P. aeruginosa strain PA14 the novel sRNA GssA. We found that it can establish a many-sided reciprocal interplay with Hfq which goes beyond the canonical mechanism of direct physical interaction that had previously been characterized for other sRNAs. Given that the Hfq-driven regulatory network of virulence factors is very broad and important for the progression of infection, we consider GssA as a new RNA target that can potentially be used to develop new antibacterial drugs.
Keywords: Hfq; Pseudomonas aeruginosa; exotoxin A; glucose utilization; posttranscriptional regulation; pyocyanin; sRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures