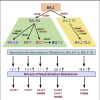
Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2
- PMID: 36476380
- PMCID: PMC9678813
- DOI: 10.1016/j.chom.2022.11.012
Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2
Abstract
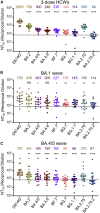
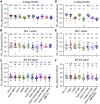
The continued evolution of SARS-CoV-2 has led to the emergence of several new Omicron subvariants, including BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Here, we examine the neutralization resistance of these subvariants against sera from 3-dose vaccinated healthcare workers, hospitalized BA.1-wave patients, and BA.4/5-wave patients. We found enhanced neutralization resistance in all new subvariants, especially in the BQ.1 and BQ.1.1 subvariants driven by N460K and K444T mutations, as well as the BA.2.75.2 subvariant driven largely by its F486S mutation. All Omicron subvariants maintained their weakened infectivity in Calu-3 cells, with the F486S mutation driving further diminished titer for the BA.2.75.2 subvariant. Molecular modeling revealed the mechanisms of antibody-mediated immune evasion by R346T, K444T, F486S, and D1199N mutations. Altogether, these findings shed light on the evolution of newly emerging SARS-CoV-2 Omicron subvariants.
Keywords: Omicron subvariants; immune evasion; molecular modeling; mutations; neutralization.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Distinct Neutralizing Antibody Escape of SARS-CoV-2 Omicron Subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2.bioRxiv [Preprint]. 2022 Oct 20:2022.10.19.512891. doi: 10.1101/2022.10.19.512891. bioRxiv. 2022. Update in: Cell Host Microbe. 2023 Jan 11;31(1):9-17.e3. doi: 10.1016/j.chom.2022.11.012. PMID: 36299423 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

