Characterization of mutant versions of the R-RAS2/TC21 GTPase found in tumors
- PMID: 36476833
- PMCID: PMC9883167
- DOI: 10.1038/s41388-022-02563-9
Characterization of mutant versions of the R-RAS2/TC21 GTPase found in tumors
Abstract
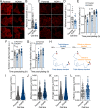
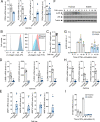
The R-RAS2 GTP hydrolase (GTPase) (also known as TC21) has been traditionally considered quite similar to classical RAS proteins at the regulatory and signaling levels. Recently, a long-tail hotspot mutation targeting the R-RAS2/TC21 Gln72 residue (Q72L) was identified as a potent oncogenic driver. Additional point mutations were also found in other tumors at low frequencies. Despite this, little information is available regarding the transforming role of these mutant versions and their relevance for the tumorigenic properties of already-transformed cancer cells. Here, we report that many of the RRAS2 mutations found in human cancers are highly transforming when expressed in immortalized cell lines. Moreover, the expression of endogenous R-RAS2Q72L is important for maintaining optimal levels of PI3K and ERK activities as well as for the adhesion, invasiveness, proliferation, and mitochondrial respiration of ovarian and breast cancer cell lines. Endogenous R-RAS2Q72L also regulates gene expression programs linked to both cell adhesion and inflammatory/immune-related responses. Endogenous R-RAS2Q72L is also quite relevant for the in vivo tumorigenic activity of these cells. This dependency is observed even though these cancer cell lines bear concurrent gain-of-function mutations in genes encoding RAS signaling elements. Finally, we show that endogenous R-RAS2, unlike the case of classical RAS proteins, specifically localizes in focal adhesions. Collectively, these results indicate that gain-of-function mutations of R-RAS2/TC21 play roles in tumor initiation and maintenance that are not fully redundant with those regulated by classical RAS oncoproteins.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, et al. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J Biol Chem. 2000;275:20020–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

