Lipid-laden lung mesenchymal cells foster breast cancer metastasis via metabolic reprogramming of tumor cells and natural killer cells
- PMID: 36476935
- PMCID: PMC9819197
- DOI: 10.1016/j.cmet.2022.11.003
Lipid-laden lung mesenchymal cells foster breast cancer metastasis via metabolic reprogramming of tumor cells and natural killer cells
Abstract
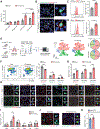
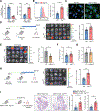
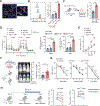
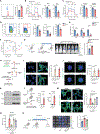
While the distant organ environment is known to support metastasis of primary tumors, its metabolic roles in this process remain underdetermined. Here, in breast cancer models, we found lung-resident mesenchymal cells (MCs) accumulating neutral lipids at the pre-metastatic stage. This was partially mediated by interleukin-1β (IL-1β)-induced hypoxia-inducible lipid droplet-associated (HILPDA) that subsequently represses adipose triglyceride lipase (ATGL) activity in lung MCs. MC-specific ablation of the ATGL or HILPDA genes in mice reinforced and reduced lung metastasis of breast cancer respectively, suggesting a metastasis-promoting effect of lipid-laden MCs. Mechanistically, lipid-laden MCs transported their lipids to tumor cells and natural killer (NK) cells via exosome-like vesicles, leading to heightened tumor cell survival and proliferation and NK cell dysfunction. Blockage of IL-1β, which was effective singly, improved the efficacy of adoptive NK cell immunotherapy in mitigating lung metastasis. Collectively, lung MCs metabolically regulate tumor cells and anti-tumor immunity to facilitate breast cancer lung metastasis.
Keywords: IL-1β; NK cells; breast cancer; disseminated tumor cells; inflammation; lipid storage; lung metastasis; mesenchymal cells; metabolic reprograming; triglycerides.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing of interests.
Figures



Comment in
-
Not so neutral lipids: Metabolic regulation of the pre-metastatic niche.Cell Metab. 2022 Dec 6;34(12):1899-1900. doi: 10.1016/j.cmet.2022.11.007. Cell Metab. 2022. PMID: 36476931 Free PMC article.
References
-
- Chaffer CL & Weinberg RA A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). - PubMed
-
- Peinado H et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer 17, 302–317 (2017). - PubMed
-
- Miranda F et al. Salt-Inducible Kinase 2 Couples Ovarian Cancer Cell Metabolism with Survival at the Adipocyte-Rich Metastatic Niche. Cancer Cell 30, 273–289 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

