CLSTN3β enforces adipocyte multilocularity to facilitate lipid utilization
- PMID: 36477540
- PMCID: PMC9995219
- DOI: 10.1038/s41586-022-05507-1
CLSTN3β enforces adipocyte multilocularity to facilitate lipid utilization
Abstract
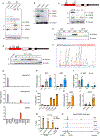
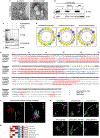
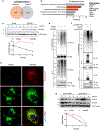
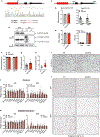
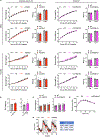
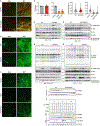
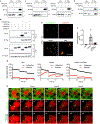
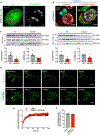
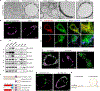
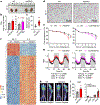
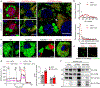
Multilocular adipocytes are a hallmark of thermogenic adipose tissue1,2, but the factors that enforce this cellular phenotype are largely unknown. Here, we show that an adipocyte-selective product of the Clstn3 locus (CLSTN3β) present in only placental mammals facilitates the efficient use of stored triglyceride by limiting lipid droplet (LD) expansion. CLSTN3β is an integral endoplasmic reticulum (ER) membrane protein that localizes to ER-LD contact sites through a conserved hairpin-like domain. Mice lacking CLSTN3β have abnormal LD morphology and altered substrate use in brown adipose tissue, and are more susceptible to cold-induced hypothermia despite having no defect in adrenergic signalling. Conversely, forced expression of CLSTN3β is sufficient to enforce a multilocular LD phenotype in cultured cells and adipose tissue. CLSTN3β associates with cell death-inducing DFFA-like effector proteins and impairs their ability to transfer lipid between LDs, thereby restricting LD fusion and expansion. Functionally, increased LD surface area in CLSTN3β-expressing adipocytes promotes engagement of the lipolytic machinery and facilitates fatty acid oxidation. In human fat, CLSTN3B is a selective marker of multilocular adipocytes. These findings define a molecular mechanism that regulates LD form and function to facilitate lipid utilization in thermogenic adipocytes.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures

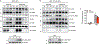



Comment in
-
Keeping brown fat brown.Sci Signal. 2022 Dec 20;15(765):eadg3025. doi: 10.1126/scisignal.adg3025. Epub 2022 Dec 20. Sci Signal. 2022. PMID: 36538591
References
-
- Cannon B & Nedergaard J Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359 (2004). - PubMed
-
- Rasmussen AT The so-called hibernating gland. J. Morphol 38, 147–205 (1923).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

