The TRAPP complexes: oligomeric exchange factors that activate the small GTPases Rab1 and Rab11
- PMID: 36477798
- PMCID: PMC10152722
- DOI: 10.1002/1873-3468.14553
The TRAPP complexes: oligomeric exchange factors that activate the small GTPases Rab1 and Rab11
Abstract
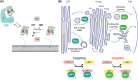
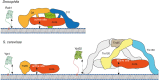
The Transport Protein Particle (TRAPP) complexes are highly conserved multisubunit complexes that act as nucleotide exchange factors (GEFs) for Rab GTPases. They act in both protein secretion and autophagy and have also been proposed to have a role in other processes such as cytokinesis and ciliogenesis. There are two TRAPP complexes in metazoans: TRAPPII, which activates Rab11; and TRAPPIII, which activates Rab1. Both complexes share a core of small subunits that form the active site for the exchange of GDP for GTP. In addition, each TRAPP complex has distinct large subunits that determine the specificity of each complex towards its substrate Rab and are essential for activity in vivo. Crystal structures have revealed the organisation of the TRAPP core and the mechanism of Rab1 activation, whilst recent cryo-EM structures have unveiled the arrangement of the specific subunits around the core to form each complex. Combining these findings with functional experiments has allowed the proposal of mechanisms for how the specificity of each complex towards their cognate Rab is determined and for the arrangement of these large complexes on the membrane.
Keywords: Golgi apparatus; Rab GTPase; autophagy; exchange factor; membrane traffic; recycling endosome.
© 2022 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures



Similar articles
-
TRAPPopathies: Severe Multisystem Disorders Caused by Variants in Genes of the Transport Protein Particle (TRAPP) Complexes.Int J Mol Sci. 2024 Dec 12;25(24):13329. doi: 10.3390/ijms252413329. Int J Mol Sci. 2024. PMID: 39769094 Free PMC article. Review.
-
Biochemical Insight into Novel Rab-GEF Activity of the Mammalian TRAPPIII Complex.J Mol Biol. 2021 Sep 3;433(18):167145. doi: 10.1016/j.jmb.2021.167145. Epub 2021 Jul 3. J Mol Biol. 2021. PMID: 34229011
-
The two TRAPP complexes of metazoans have distinct roles and act on different Rab GTPases.J Cell Biol. 2018 Feb 5;217(2):601-617. doi: 10.1083/jcb.201705068. Epub 2017 Dec 22. J Cell Biol. 2018. PMID: 29273580 Free PMC article.
-
The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway.J Cell Biol. 2018 Jan 2;217(1):283-298. doi: 10.1083/jcb.201705214. Epub 2017 Nov 6. J Cell Biol. 2018. PMID: 29109089 Free PMC article.
-
The TRAPP complexes: discriminating GTPases in context.FEBS Lett. 2023 Mar;597(6):721-733. doi: 10.1002/1873-3468.14557. Epub 2022 Dec 21. FEBS Lett. 2023. PMID: 36481981 Free PMC article. Review.
Cited by
-
Biochemical Structure and Function of TRAPP Complexes in the Cardiac System.JACC Basic Transl Sci. 2023 Jul 12;8(12):1599-1612. doi: 10.1016/j.jacbts.2023.03.011. eCollection 2023 Dec. JACC Basic Transl Sci. 2023. PMID: 38205348 Free PMC article.
-
Structural insights into traffic through the Golgi complex.Curr Opin Cell Biol. 2025 Jun;94:102505. doi: 10.1016/j.ceb.2025.102505. Epub 2025 Mar 28. Curr Opin Cell Biol. 2025. PMID: 40157309 Review.
-
TRAPPopathies: Severe Multisystem Disorders Caused by Variants in Genes of the Transport Protein Particle (TRAPP) Complexes.Int J Mol Sci. 2024 Dec 12;25(24):13329. doi: 10.3390/ijms252413329. Int J Mol Sci. 2024. PMID: 39769094 Free PMC article. Review.
-
PhyloString: A web server designed to identify, visualize, and evaluate functional relationships between orthologous protein groups across different phylogenetic lineages.PLoS One. 2024 Jan 26;19(1):e0297010. doi: 10.1371/journal.pone.0297010. eCollection 2024. PLoS One. 2024. PMID: 38277370 Free PMC article.
-
Unveiling the TRAPP: The role of plant TRAPPII in adaptive growth decisions.J Cell Biol. 2024 May 6;223(5):e202404039. doi: 10.1083/jcb.202404039. Epub 2024 Apr 23. J Cell Biol. 2024. PMID: 38652246 Free PMC article.
References
-
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. - PubMed
-
- Muschalik N, Munro S. Golgins. Curr Biol. 2018;28:R374–6. - PubMed
-
- Ungermann C, Kummel D. Structure of membrane tethers and their role in fusion. Traffic. 2019;20:479–90. - PubMed
-
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. - PubMed
-
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

