Hijacking the human complement inhibitor C4b-binding protein by the sporozoite stage of the Plasmodium falciparum parasite
- PMID: 36479121
- PMCID: PMC9720182
- DOI: 10.3389/fimmu.2022.1051161
Hijacking the human complement inhibitor C4b-binding protein by the sporozoite stage of the Plasmodium falciparum parasite
Abstract
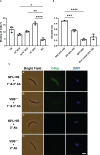
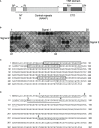
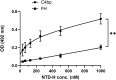
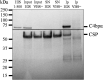
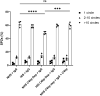
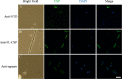
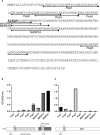
The complement system is considered the first line of defense against pathogens. Hijacking complement regulators from blood is a common evasion tactic of pathogens to inhibit complement activation on their surfaces. Here, we report hijacking of the complement C4b-binding protein (C4bp), the regulator of the classical and lectin pathways of complement activation, by the sporozoite (SPZ) stage of the Plasmodium falciparum parasite. This was shown by direct binding of radiolabeled purified C4bp to live SPZs as well as by binding of C4bp from human serum to SPZs in indirect immunofluorescence assays. Using a membrane-bound peptide array, peptides from the N-terminal domain (NTD) of P. falciparum circumsporozoite protein (CSP) were found to bind C4bp. Soluble biotinylated peptide covering the same region on the NTD and a recombinantly expressed NTD also bound C4bp in a dose-dependent manner. NTD-binding site on C4bp was mapped to the CCP1-2 of the C4bp α-chain, a common binding site for many pathogens. Native CSP was also co-immunoprecipitated with C4bp from human serum. Preventing C4bp binding to the SPZ surface negatively affected the SPZs gliding motility in the presence of functional complement and malaria hyperimmune IgG confirming the protective role of C4bp in controlling complement activation through the classical pathway on the SPZ surface. Incorporating the CSP-C4bp binding region into a CSP-based vaccine formulation could induce vaccine-mediated immunity that neutralizes this immune evasion region and increases the vaccine efficacy.
Keywords: C4b binding protein; Plasmodium; circumsporozoite protein; complement evasion; sporozoites.
Copyright © 2022 Khattab, Rezola, Barroso, Kyrklund, Pihlajamaa, Freitag, van Gemert, Bousema, Permi, Turunen, Sauerwein, Luty and Meri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

