Potential role of microRNAs in selective hepatic insulin resistance: From paradox to the paradigm
- PMID: 36479211
- PMCID: PMC9720316
- DOI: 10.3389/fendo.2022.1028846
Potential role of microRNAs in selective hepatic insulin resistance: From paradox to the paradigm
Abstract
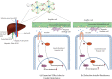
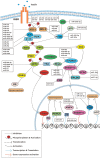
The paradoxical action of insulin on hepatic glucose metabolism and lipid metabolism in the insulin-resistant state has been of much research interest in recent years. Generally, insulin resistance would promote hepatic gluconeogenesis and demote hepatic de novo lipogenesis. The underlying major drivers of these mechanisms were insulin-dependent, via FOXO-1-mediated gluconeogenesis and SREBP1c-mediated lipogenesis. However, insulin-resistant mouse models have shown high glucose levels as well as excess lipid accumulation. As suggested, the inert insulin resistance causes the activation of the FOXO-1 pathway promoting gluconeogenesis. However, it does not affect the SREBP1c pathway; therefore, cells continue de novo lipogenesis. Many hypotheses were suggested for this paradoxical action occurring in insulin-resistant rodent models. A "downstream branch point" in the insulin-mediated pathway was suggested to act differentially on the FOXO-1 and SREBP1c pathways. MicroRNAs have been widely studied for their action of pathway mediation via suppressing the intermediate protein expressions. Many in vitro studies have postulated the roles of hepato-specific expressions of miRNAs on insulin cascade. Thus, miRNA would play a pivotal role in selective hepatic insulin resistance. As observed, there were confirmations and contradictions between the outcomes of gene knockout studies conducted on selective hepatic insulin resistance and hepato-specific miRNA expression studies. Furthermore, these studies had evaluated only the effect of miRNAs on glucose metabolism and few on hepatic de novo lipogenesis, limiting the ability to conclude their role in selective hepatic insulin resistance. Future studies conducted on the role of miRNAs on selective hepatic insulin resistance warrant the understanding of this paradoxical action of insulin.
Keywords: FOXO-1; SREBP1c; microRNA; paradox; role; selective hepatic insulin resistance.
Copyright © 2022 Palihaderu, Mendis, Premarathne, Dias, Yeap, Ho, Dissanayake, Rajapakse, Karunanayake, Senarath and Satharasinghe.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ozougwu O. The pathogenesis and pathophysiology of type 1 and type 2 diabetes mellitus. J Physiol Pathophysiol (2013) 4(4):46–57. doi: 10.5897/JPAP2013.0001 - DOI
-
- Williams R, Colagiuri S, Chan J, Gregg EW, Ke C, Lim L, et al. . IDF diabetes atlas. 9th edition. Brussels, Belgium: International Diabetes Federation; (2019). Available at: https://www.diabetesatlas.org/en/.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

