Genomic profiling and network-level understanding uncover the potential genes and the pathways in hepatocellular carcinoma
- PMID: 36479247
- PMCID: PMC9720179
- DOI: 10.3389/fgene.2022.880440
Genomic profiling and network-level understanding uncover the potential genes and the pathways in hepatocellular carcinoma
Abstract
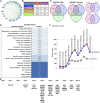
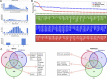
Data integration with phenotypes such as gene expression, pathways or function, and protein-protein interactions data has proven to be a highly promising technique for improving human complex diseases, particularly cancer patient outcome prediction. Hepatocellular carcinoma is one of the most prevalent cancers, and the most common cause is chronic HBV and HCV infection, which is linked to the majority of cases, and HBV and HCV play a role in multistep carcinogenesis progression. We examined the list of known hepatocellular carcinoma biomarkers with the publicly available expression profile dataset of hepatocellular carcinoma infected with HCV from day 1 to day 10 in this study. The study covers an overexpression pattern for the selected biomarkers in clinical hepatocellular carcinoma patients, a combined investigation of these biomarkers with the gathered temporal dataset, temporal expression profiling changes, and temporal pathway enrichment following HCV infection. Following a temporal analysis, it was discovered that the early stages of HCV infection tend to be more harmful in terms of expression shifting patterns, and that there is no significant change after that, followed by a set of genes that are consistently altered. PI3K, cAMP, TGF, TNF, Rap1, NF-kB, Apoptosis, Longevity regulating pathway, signaling pathways regulating pluripotency of stem cells, Cytokine-cytokine receptor interaction, p53 signaling, Wnt signaling, Toll-like receptor signaling, and Hippo signaling pathways are just a few of the most commonly enriched pathways. The majority of these pathways are well-known for their roles in the immune system, infection and inflammation, and human illnesses like cancer. We also find that ADCY8, MYC, PTK2, CTNNB1, TP53, RB1, PRKCA, TCF7L2, PAK1, ITPR2, CYP3A4, UGT1A6, GCK, and FGFR2/3 appear to be among the prominent genes based on the networks of genes and pathways based on the copy number alterations, mutations, and structural variants study.
Keywords: HCV and HCC; biomarkers; co-expression; gene expression/mutational profiling; network-level understanding.
Copyright © 2022 El-Kafrawy, El-Daly, Bajrai, Alandijany, Faizo, Mobashir, Ahmed, Ahmed, Alam, Jeet, Kamal, Anwer, Khan, Tashkandi, Rizvi and Azhar.
Conflict of interest statement
Author MK was employed by the company Enzymoics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Role of Potential COVID-19 Immune System Associated Genes and the Potential Pathways Linkage with Type-2 Diabetes.Comb Chem High Throughput Screen. 2022;25(14):2452-2462. doi: 10.2174/1386207324666210804124416. Comb Chem High Throughput Screen. 2022. PMID: 34348612
-
Molecular cytogenetic evaluation of virus-associated and non-viral hepatocellular carcinoma: analysis of 26 carcinomas and 12 concurrent dysplasias.J Pathol. 2000 Oct;192(2):207-15. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH690>3.0.CO;2-#. J Pathol. 2000. PMID: 11004697
-
A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma.BMC Med Genomics. 2019 Oct 28;12(1):147. doi: 10.1186/s12920-019-0580-x. BMC Med Genomics. 2019. PMID: 31660973 Free PMC article.
-
Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections.Genomics. 2013 Aug;102(2):74-83. doi: 10.1016/j.ygeno.2013.04.001. Epub 2013 Apr 11. Genomics. 2013. PMID: 23583669 Review.
-
Epigenetic Mechanisms Involved in HCV-Induced Hepatocellular Carcinoma (HCC).Front Oncol. 2021 Jul 15;11:677926. doi: 10.3389/fonc.2021.677926. eCollection 2021. Front Oncol. 2021. PMID: 34336665 Free PMC article. Review.
Cited by
-
Unlocking vinpocetine's oncostatic potential in early-stage hepatocellular carcinoma: A new approach to oncogenic modulation by a nootropic drug.PLoS One. 2024 Oct 31;19(10):e0312572. doi: 10.1371/journal.pone.0312572. eCollection 2024. PLoS One. 2024. PMID: 39480853 Free PMC article.
-
Development and evaluation of a risk score model based on a WNT score gene-associated signature for predicting the clinical outcome and the tumour microenvironment of hepatocellular carcinoma.Int J Immunopathol Pharmacol. 2023 Jan-Dec;37:3946320231218179. doi: 10.1177/03946320231218179. Int J Immunopathol Pharmacol. 2023. PMID: 38054921 Free PMC article.
-
A comparison of hepatitis B virus- and hepatitis C virus-related hepatocellular carcinoma: a bioinformatics analysis.Transl Cancer Res. 2025 Jul 30;14(7):4243-4259. doi: 10.21037/tcr-2024-2607. Epub 2025 Jul 23. Transl Cancer Res. 2025. PMID: 40792161 Free PMC article.
-
Characterization of the Rat Osteosarcoma Cell Line UMR-106 by Long-Read Technologies Identifies a Large Block of Amplified Genes Associated with Human Disease.Genes (Basel). 2024 Sep 26;15(10):1254. doi: 10.3390/genes15101254. Genes (Basel). 2024. PMID: 39457378 Free PMC article.
-
NF-kB in Signaling Patterns and Its Temporal Dynamics Encode/Decode Human Diseases.Life (Basel). 2022 Dec 2;12(12):2012. doi: 10.3390/life12122012. Life (Basel). 2022. PMID: 36556376 Free PMC article. Review.
References
-
- Agell L., Hernández S., Nonell L., Lorenzo M., Puigdecanet E., de Muga S., et al. (2012). A 12-gene expression signature is associated with aggressive histological in prostate cancer: SEC14L1 and TCEB1 genes are potential markers of progression. Am. J. Pathol. 181, 1585–1594. 10.1016/j.ajpath.2012.08.005 - DOI - PubMed
-
- Anwer S. T., Mobashir M., Fantoukh O. I., Khan B., Imtiyaz K., Naqvi I. H., et al. (2022). Synthesis of silver nano particles using myricetin and the in-vitro assessment of anti-colorectal cancer activity: In-silico integration. Int. J. Mol. Sci. 23, 11024–11118. 10.3390/ijms231911024 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

