A yeast-based tool for screening mammalian diacylglycerol acyltransferase inhibitors
- PMID: 36479627
- PMCID: PMC9716225
- DOI: 10.1002/mbo3.1334
A yeast-based tool for screening mammalian diacylglycerol acyltransferase inhibitors
Abstract
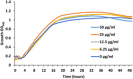
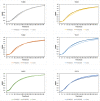
Dysregulation of lipid metabolism is associated with obesity and metabolic diseases but there is also increasing evidence of a relationship between lipid body excess and cancer. Lipid body synthesis requires diacylglycerol acyltransferases (DGATs) which catalyze the last step of triacylglycerol synthesis from diacylglycerol and acyl-coenzyme A. The DGATs and in particular DGAT2, are therefore considered potential therapeutic targets for the control of these pathologies. Here, the murine and the human DGAT2 were overexpressed in the oleaginous yeast Yarrowia lipolytica deleted for all DGAT activities, to evaluate the functionality of the enzymes in this heterologous host and DGAT activity inhibitors. This work provides evidence that mammalian DGATs expressed in Y. lipolytica are a useful tool for screening chemical libraries to identify potential inhibitors or activators of these enzymes of therapeutic interest.
Keywords: DGAT; Yarrowia lipolytica; acyl-CoA:diacylglycerol acyltransferase; heterologous expression; inhibitor screening.
© 2022 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures







References
-
- Almanza, A. , Mnich, K. , Blomme, A. , Robinson, C. M. , Rodriguez‐Blanco, G. , Kierszniowska, S. , McGrath, E. P. , Le Gallo, M. , Pilalis, E. , Swinnen, J. V. , Chatziioannou, A. , Chevet, E. , Gorman, A. M. , & Samali, A. (2022). Regulated IRE1α‐dependent decay (RIDD)‐mediated reprograming of lipid metabolism in cancer. Nature Communications, 13, 2493. - PMC - PubMed
-
- Beopoulos, A. , Haddouche, R. , Kabran, P. , Dulermo, T. , Chardot, T. , & Nicaud, J.‐M. (2012). Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl‐CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. new insights into the storage lipid metabolism of oleaginous yeasts. Applied Microbiology and Biotechnology, 93, 1523–1537. - PMC - PubMed
-
- Browse, J. , McCourt, P. J. , & Somerville, C. R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Analytical Biochemistry, 152, 141–145. - PubMed
-
- Cases, S. , Smith, S. J. , Zheng, Y.‐W. , Myers, H. M. , Lear, S. R. , Sande, E. , Novak, S. , Collins, C. , Welch, C. B. , Lusis, A. J. , Erickson, S. K. , & Farese, R. V. (1998). Identification of a gene encoding an acyl CoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proceedings of the National Academy of Sciences USA, 95, 13018–13023. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

