Denisovan introgression has shaped the immune system of present-day Papuans
- PMID: 36480515
- PMCID: PMC9731433
- DOI: 10.1371/journal.pgen.1010470
Denisovan introgression has shaped the immune system of present-day Papuans
Abstract
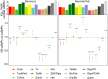
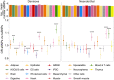
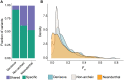
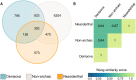
Modern humans have admixed with multiple archaic hominins. Papuans, in particular, owe up to 5% of their genome to Denisovans, a sister group to Neanderthals whose remains have only been identified in Siberia and Tibet. Unfortunately, the biological and evolutionary significance of these introgression events remain poorly understood. Here we investigate the function of both Denisovan and Neanderthal alleles characterised within a set of 56 genomes from Papuan individuals. By comparing the distribution of archaic and non-archaic variants we assess the consequences of archaic admixture across a multitude of different cell types and functional elements. We observe an enrichment of archaic alleles within cis-regulatory elements and transcribed regions of the genome, with Denisovan variants strongly affecting elements active within immune-related cells. We identify 16,048 and 10,032 high-confidence Denisovan and Neanderthal variants that fall within annotated cis-regulatory elements and with the potential to alter the affinity of multiple transcription factors to their cognate DNA motifs, highlighting a likely mechanism by which introgressed DNA can impact phenotypes. Lastly, we experimentally validate these predictions by testing the regulatory potential of five Denisovan variants segregating within Papuan individuals, and find that two are associated with a significant reduction of transcriptional activity in plasmid reporter assays. Together, these data provide support for a widespread contribution of archaic DNA in shaping the present levels of modern human genetic diversity, with different archaic ancestries potentially affecting multiple phenotypic traits within non-Africans.
Copyright: © 2022 Vespasiani et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Lorente-Galdos B, Lao O, Serra-Vidal G, Santpere G, Kuderna LFK, Arauna LR, et al.. Whole-genome sequence analysis of a Pan African set of samples reveals archaic gene flow from an extinct basal population of modern humans into sub-Saharan populations. Genome Biology. 2019;20(1):1–15. doi: 10.1186/s13059-019-1684-5 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

