LDAK-GBAT: Fast and powerful gene-based association testing using summary statistics
- PMID: 36480927
- PMCID: PMC9892699
- DOI: 10.1016/j.ajhg.2022.11.010
LDAK-GBAT: Fast and powerful gene-based association testing using summary statistics
Abstract
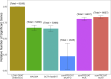
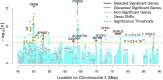
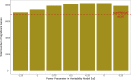
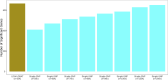
We present LDAK-GBAT, a tool for gene-based association testing using summary statistics from genome-wide association studies that is computationally efficient, produces well-calibrated p values, and is significantly more powerful than existing tools. LDAK-GBAT takes approximately 30 min to analyze imputed data (2.9M common, genic SNPs), requiring less than 10 Gb memory. It shows good control of type 1 error given an appropriate reference panel. Across 109 phenotypes (82 from the UK Biobank, 18 from the Million Veteran Program, and nine from the Psychiatric Genetics Consortium), LDAK-GBAT finds on average 19% (SE: 1%) more significant genes than the existing tool sumFREGAT-ACAT, with even greater gains in comparison with MAGMA, GCTA-fastBAT, sumFREGAT-SKAT-O, and sumFREGAT-PCA.
Keywords: UK Biobank; complex traits; gene-based association testing; genome-wide association study; statistical genetics.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

