Host microRNAs exhibit differential propensity to interact with SARS-CoV-2 and variants of concern
- PMID: 36481486
- PMCID: PMC9721271
- DOI: 10.1016/j.bbadis.2022.166612
Host microRNAs exhibit differential propensity to interact with SARS-CoV-2 and variants of concern
Abstract
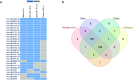
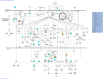
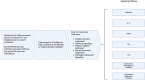
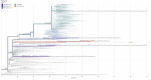
A significant number of SARS-CoV-2-infected individuals naturally overcome viral infection, suggesting the existence of a potent endogenous antiviral mechanism. As an innate defense mechanism, microRNA (miRNA) pathways in mammals have evolved to restrict viruses, besides regulating endogenous mRNAs. In this study, we systematically examined the complete repertoire of human miRNAs for potential binding sites on SARS-CoV-2 Wuhan-Hu-1, Beta, Delta, and Omicron. Human miRNA and viral genome interaction were analyzed using RNAhybrid 2.2 with stringent parameters to identify highly bonafide miRNA targets. Using publicly available data, we filtered for miRNAs expressed in lung epithelial cells/tissue and oral keratinocytes, concentrating on the miRNAs that target SARS-CoV-2 S protein mRNAs. Our results show a significant loss of human miRNA and SARS-CoV-2 interactions in Omicron (130 miRNAs) compared to Wuhan-Hu-1 (271 miRNAs), Beta (279 miRNAs), and Delta (275 miRNAs). In particular, hsa-miR-3150b-3p and hsa-miR-4784 show binding affinity for S protein of Wuhan strain but not Beta, Delta, and Omicron. Loss of miRNA binding sites on N protein was also observed for Omicron. Through Ingenuity Pathway Analysis (IPA), we examined the experimentally validated and highly predicted functional role of these miRNAs. We found that hsa-miR-3150b-3p and hsa-miR-4784 have several experimentally validated or highly predicted target genes in the Toll-like receptor, IL-17, Th1, Th2, interferon, and coronavirus pathogenesis pathways. Focusing on the coronavirus pathogenesis pathway, we found that hsa-miR-3150b-3p and hsa-miR-4784 are highly predicted to target MAPK13. Exploring miRNAs to manipulate viral genome/gene expression can provide a promising strategy with successful outcomes by targeting specific VOCs.
Keywords: Antiviral immunity; MicroRNAs; Post-transcriptional regulation; SARS-CoV-2; Variants of concern.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
SARS-COV-2 as potential microRNA sponge in COVID-19 patients.BMC Med Genomics. 2022 Apr 23;15(Suppl 2):94. doi: 10.1186/s12920-022-01243-7. BMC Med Genomics. 2022. PMID: 35461273 Free PMC article.
-
Computational Analysis of Targeting SARS-CoV-2, Viral Entry Proteins ACE2 and TMPRSS2, and Interferon Genes by Host MicroRNAs.Genes (Basel). 2020 Nov 16;11(11):1354. doi: 10.3390/genes11111354. Genes (Basel). 2020. PMID: 33207533 Free PMC article.
-
Comparative study of predicted miRNA between Indonesia and China (Wuhan) SARS-CoV-2: a bioinformatics analysis.Genes Genomics. 2021 Sep;43(9):1079-1086. doi: 10.1007/s13258-021-01119-7. Epub 2021 Jun 21. Genes Genomics. 2021. PMID: 34152577 Free PMC article.
-
The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review.Infect Genet Evol. 2021 Jul;91:104832. doi: 10.1016/j.meegid.2021.104832. Epub 2021 Apr 1. Infect Genet Evol. 2021. PMID: 33812037 Free PMC article.
-
MicroRNAs as Potential Tools for Predicting Cancer Patients' Susceptibility to SARS-CoV-2 Infection and Vaccination Response.Cells. 2022 Jul 23;11(15):2279. doi: 10.3390/cells11152279. Cells. 2022. PMID: 35892576 Free PMC article. Review.
Cited by
-
Decoding the intricacies: a comprehensive analysis of microRNAs in the pathogenesis, diagnosis, prognosis and therapeutic strategies for COVID-19.Front Med (Lausanne). 2024 Oct 7;11:1430974. doi: 10.3389/fmed.2024.1430974. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39434774 Free PMC article. Review.
-
Endogenous miRNA-Based Innate-Immunity against SARS-CoV-2 Invasion of the Brain.Int J Mol Sci. 2023 Feb 8;24(4):3363. doi: 10.3390/ijms24043363. Int J Mol Sci. 2023. PMID: 36834773 Free PMC article. Review.
-
Oral SARS-CoV-2 Infection and Risk for Long Covid.Rev Med Virol. 2025 Mar;35(2):e70029. doi: 10.1002/rmv.70029. Rev Med Virol. 2025. PMID: 40074704 Free PMC article. Review.
References
-
- WHO Tracking SARS-CoV-2 variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

