The Liver Tumor Segmentation Benchmark (LiTS)
- PMID: 36481607
- PMCID: PMC10631490
- DOI: 10.1016/j.media.2022.102680
The Liver Tumor Segmentation Benchmark (LiTS)
Abstract
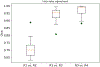
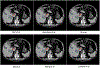
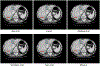
In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LiTS), which was organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2017 and the International Conferences on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2017 and 2018. The image dataset is diverse and contains primary and secondary tumors with varied sizes and appearances with various lesion-to-background levels (hyper-/hypo-dense), created in collaboration with seven hospitals and research institutions. Seventy-five submitted liver and liver tumor segmentation algorithms were trained on a set of 131 computed tomography (CT) volumes and were tested on 70 unseen test images acquired from different patients. We found that not a single algorithm performed best for both liver and liver tumors in the three events. The best liver segmentation algorithm achieved a Dice score of 0.963, whereas, for tumor segmentation, the best algorithms achieved Dices scores of 0.674 (ISBI 2017), 0.702 (MICCAI 2017), and 0.739 (MICCAI 2018). Retrospectively, we performed additional analysis on liver tumor detection and revealed that not all top-performing segmentation algorithms worked well for tumor detection. The best liver tumor detection method achieved a lesion-wise recall of 0.458 (ISBI 2017), 0.515 (MICCAI 2017), and 0.554 (MICCAI 2018), indicating the need for further research. LiTS remains an active benchmark and resource for research, e.g., contributing the liver-related segmentation tasks in http://medicaldecathlon.com/. In addition, both data and online evaluation are accessible via https://competitions.codalab.org/competitions/17094.
Keywords: Benchmark; CT; Deep learning; Liver; Liver tumor; Segmentation.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







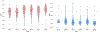

References
-
- Abdel-massieh NH, Hadhoud MM, Amin KM, 2010. Fully automatic liver tumor segmentation from abdominal CT scans. In: Computer Engineering and Systems (ICCES), 2010 International Conference on. IEEE, pp. 197–202.
-
- Albain KS, Swann RS, Rusch VW, Turrisi AT, Shepherd FA, Smith C, Chen Y, Livingston RB, Feins RH, Gandara DR, et al., 2009. Radiotherapy plus chemotherapy with or without surgical resection for stage III non-small-cell lung cancer: A phase III randomised controlled trial. Lancet 374 (9687), 379–386. - PMC - PubMed
-
- Amiranashvili T, Lüdke D, Li H, Zachow S, et al., 2021. Learning shape reconstruction from sparse measurements with neural implicit functions. In: Medical Imaging with Deep Learning.
-
- Bauknecht H-C, Romano VC, Rogalla P, Klingebiel R, Wolf C, Bornemann L, Hamm B, Hein PA, 2010. Intra-and interobserver variability of linear and volumetric measurements of brain metastases using contrast-enhanced magnetic resonance imaging. Invest. Radiol 45 (1), 49–56. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

