Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae
- PMID: 36482101
- PMCID: PMC9873574
- DOI: 10.1038/s41591-022-02107-4
Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae
Abstract
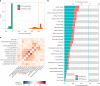
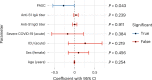
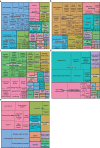
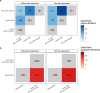
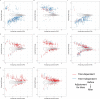
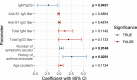
Post-acute sequelae of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are debilitating, clinically heterogeneous and of unknown molecular etiology. A transcriptome-wide investigation was performed in 165 acutely infected hospitalized individuals who were followed clinically into the post-acute period. Distinct gene expression signatures of post-acute sequelae were already present in whole blood during acute infection, with innate and adaptive immune cells implicated in different symptoms. Two clusters of sequelae exhibited divergent plasma-cell-associated gene expression patterns. In one cluster, sequelae associated with higher expression of immunoglobulin-related genes in an anti-spike antibody titer-dependent manner. In the other, sequelae associated independently of these titers with lower expression of immunoglobulin-related genes, indicating lower non-specific antibody production in individuals with these sequelae. This relationship between lower total immunoglobulins and sequelae was validated in an external cohort. Altogether, multiple etiologies of post-acute sequelae were already detectable during SARS-CoV-2 infection, directly linking these sequelae with the acute host response to the virus and providing early insights into their development.
© 2022. The Author(s).
Conflict of interest statement
S.G. reports other research funding from Regeneron, Genentech, Boehringer Ingelheim, EMD Serono, Takeda, Bristol Myers Squibb and Celgene. The remaining authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

