A critical period of translational control during brain development at codon resolution
- PMID: 36482253
- PMCID: PMC9758057
- DOI: 10.1038/s41594-022-00882-9
A critical period of translational control during brain development at codon resolution
Erratum in
-
Author Correction: A critical period of translational control during brain development at codon resolution.Nat Struct Mol Biol. 2025 Sep;32(9):1838. doi: 10.1038/s41594-025-01675-6. Nat Struct Mol Biol. 2025. PMID: 40866578 Free PMC article. No abstract available.
Abstract
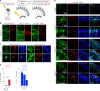
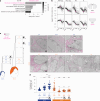
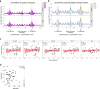
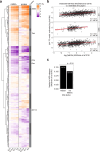
Translation modulates the timing and amplification of gene expression after transcription. Brain development requires uniquely complex gene expression patterns, but large-scale measurements of translation directly in the prenatal brain are lacking. We measure the reactants, synthesis and products of mRNA translation spanning mouse neocortex neurogenesis, and discover a transient window of dynamic regulation at mid-gestation. Timed translation upregulation of chromatin-binding proteins like Satb2, which is essential for neuronal subtype differentiation, restricts protein expression in neuronal lineages despite broad transcriptional priming in progenitors. In contrast, translation downregulation of ribosomal proteins sharply decreases ribosome biogenesis, coinciding with a major shift in protein synthesis dynamics at mid-gestation. Changing activity of eIF4EBP1, a direct inhibitor of ribosome biogenesis, is concurrent with ribosome downregulation and affects neurogenesis of the Satb2 lineage. Thus, the molecular logic of brain development includes the refinement of transcriptional programs by translation. Modeling of the developmental neocortex translatome is provided as an open-source searchable resource at https://shiny.mdc-berlin.de/cortexomics .
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Buccitelli, C. & Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet.21, 630–644 (2020). - PubMed
-
- Telley, L. et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science364, eaav2522 (2019). - PubMed
-
- Zahr, S. K. et al. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron97, 520–537.e6 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

