Transcriptome analysis and cytochrome P450 monooxygenase reveal the molecular mechanism of Bisphenol A degradation by Pseudomonas putida strain YC-AE1
- PMID: 36482332
- PMCID: PMC9733184
- DOI: 10.1186/s12866-022-02689-6
Transcriptome analysis and cytochrome P450 monooxygenase reveal the molecular mechanism of Bisphenol A degradation by Pseudomonas putida strain YC-AE1
Abstract
Background: Bisphenol A (BPA) is a rapid spreading organic pollutant that widely used in many industries especially as a plasticizer in polycarbonate plastic and epoxy resins. BPA reported as a prominent endocrine disruptor compound that possesses estrogenic activity and fulminant toxicity. Pseudomonas putida YC-AE1 was isolated in our previous study and exerted a strong degradation capacity toward BPA at high concentrations; however, the molecular degradation mechanism is still enigmatic.
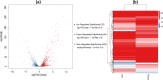
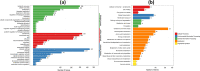
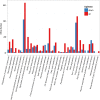
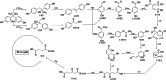
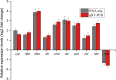
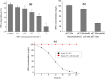
Results: We employed RNA sequencing to analyze the differentially expressed genes (DEGs) in the YC-AE1 strain upon BPA induction. Out of 1229 differentially expressed genes, 725 genes were positively regulated, and 504 genes were down-regulated. The pathways of microbial metabolism in diverse environments were significantly enriched among DEGs based on KEGG enrichment analysis. qRT-PCR confirm the involvement of BPA degradation relevant genes in accordance with RNA Seq data. The degradation pathway of BPA in YC-AE1 was proposed with specific enzymes and encoded genes. The role of cytochrome P450 (CYP450) in BPA degradation was further verified. Sever decrease in BPA degradation was recorded by YC-AE1 in the presence of CYP450 inhibitor. Subsequently, CYP450bisdB deficient YC-AE1 strain △ bisdB lost its ability toward BPA transformation comparing with the wild type. Furthermore, Transformation of E. coli with pET-32a-bisdAB empowers it to degrade 66 mg l-1 of BPA after 24 h. Altogether, the results showed the role of CYP450 in biodegradation of BPA by YC-AE1.
Conclusion: In this study we propose the molecular basis and the potential role of YC-AE1cytochrome P450 monooxygenase in BPA catabolism.
Keywords: Bisphenol A; Cytochrome P450; Degradation pathway; Pseudomonas putida YC-AE1; RNA sequencing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Dodds EC, Lawson W. Synthetic strogenic agents without the phenanthrene nucleus. Nature. 1936;137:996. doi: 10.1038/137996a0. - DOI
-
- Carlisle J, Chan D, Golub M, Henkel S, Painter P, Wu KL. Toxicological profile for bisphenol A. Integrated risk assessment branch Office of environmental health hazard assessment. Calif Environ Prot Agency. 2009;1:1–66
Publication types
MeSH terms
Substances
Grants and funding
- 31540067/National Natural Science Foundation of China
- 21876201/National Natural Science Foundation of China
- 1610042017001/Basic Research Fund of Chinese Academy of Agricultural Sciences
- 1610042018005/Basic Research Fund of Chinese Academy of Agricultural Sciences
- 1610042018006/Basic Research Fund of Chinese Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources
Miscellaneous

