De novo emergence, existence, and demise of a protein-coding gene in murids
- PMID: 36482406
- PMCID: PMC9733328
- DOI: 10.1186/s12915-022-01470-5
De novo emergence, existence, and demise of a protein-coding gene in murids
Abstract
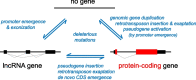
Background: Genes, principal units of genetic information, vary in complexity and evolutionary history. Less-complex genes (e.g., long non-coding RNA (lncRNA) expressing genes) readily emerge de novo from non-genic sequences and have high evolutionary turnover. Genesis of a gene may be facilitated by adoption of functional genic sequences from retrotransposon insertions. However, protein-coding sequences in extant genomes rarely lack any connection to an ancestral protein-coding sequence.
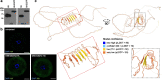
Results: We describe remarkable evolution of the murine gene D6Ertd527e and its orthologs in the rodent Muroidea superfamily. The D6Ertd527e emerged in a common ancestor of mice and hamsters most likely as a lncRNA-expressing gene. A major contributing factor was a long terminal repeat (LTR) retrotransposon insertion carrying an oocyte-specific promoter and a 5' terminal exon of the gene. The gene survived as an oocyte-specific lncRNA in several extant rodents while in some others the gene or its expression were lost. In the ancestral lineage of Mus musculus, the gene acquired protein-coding capacity where the bulk of the coding sequence formed through CAG (AGC) trinucleotide repeat expansion and duplications. These events generated a cytoplasmic serine-rich maternal protein. Knock-out of D6Ertd527e in mice has a small but detectable effect on fertility and the maternal transcriptome.
Conclusions: While this evolving gene is not showing a clear function in laboratory mice, its documented evolutionary history in Muroidea during the last ~ 40 million years provides a textbook example of how a several common mutation events can support de novo gene formation, evolution of protein-coding capacity, as well as gene's demise.
Keywords: CAG; D6Ertd527e; De novo; Evolution; Gene; LTR; Oocyte; Polyserine; Retrotransposon.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Emergence and evolution of Zfp36l3.Mol Phylogenet Evol. 2016 Jan;94(Pt B):518-530. doi: 10.1016/j.ympev.2015.10.016. Epub 2015 Oct 19. Mol Phylogenet Evol. 2016. PMID: 26493225 Free PMC article.
-
Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes.Genome Res. 2018 Apr;28(4):448-459. doi: 10.1101/gr.234096.117. Epub 2018 Mar 21. Genome Res. 2018. PMID: 29563166 Free PMC article.
-
Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes.Genome Res. 2017 Aug;27(8):1384-1394. doi: 10.1101/gr.216150.116. Epub 2017 May 18. Genome Res. 2017. PMID: 28522611 Free PMC article.
-
Retrotransposon-associated long non-coding RNAs in mice and men.Pflugers Arch. 2016 Jun;468(6):1049-60. doi: 10.1007/s00424-016-1818-5. Epub 2016 Apr 5. Pflugers Arch. 2016. PMID: 27044413 Review.
-
Fact or fiction: updates on how protein-coding genes might emerge de novo from previously non-coding DNA.F1000Res. 2017 Jan 19;6:57. doi: 10.12688/f1000research.10079.1. eCollection 2017. F1000Res. 2017. PMID: 28163910 Free PMC article. Review.
References
-
- Johannsen W. Elemente der exakten erblichkeitslehre. Deutsche wesentlich erweiterte ausgabe in fünfundzwanzig vorlesungen. Jena: G. Fischer; 1909. p. 534. https://www.archive.org/download/elementederexakt00joha/page/n4_w509.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

