Nuclear import receptors are recruited by FG-nucleoporins to rescue hallmarks of TDP-43 proteinopathy
- PMID: 36482422
- PMCID: PMC9733332
- DOI: 10.1186/s13024-022-00585-1
Nuclear import receptors are recruited by FG-nucleoporins to rescue hallmarks of TDP-43 proteinopathy
Abstract
Background: Cytoplasmic mislocalization and aggregation of TAR DNA-binding protein-43 (TDP-43) is a hallmark of the amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD) disease spectrum, causing both nuclear loss-of-function and cytoplasmic toxic gain-of-function phenotypes. While TDP-43 proteinopathy has been associated with defects in nucleocytoplasmic transport, this process is still poorly understood. Here we study the role of karyopherin-β1 (KPNB1) and other nuclear import receptors in regulating TDP-43 pathology.
Methods: We used immunostaining, immunoprecipitation, biochemical and toxicity assays in cell lines, primary neuron and organotypic mouse brain slice cultures, to determine the impact of KPNB1 on the solubility, localization, and toxicity of pathological TDP-43 constructs. Postmortem patient brain and spinal cord tissue was stained to assess KPNB1 colocalization with TDP-43 inclusions. Turbidity assays were employed to study the dissolution and prevention of aggregation of recombinant TDP-43 fibrils in vitro. Fly models of TDP-43 proteinopathy were used to determine the effect of KPNB1 on their neurodegenerative phenotype in vivo.
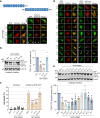
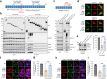
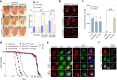
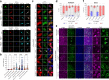
Results: We discovered that several members of the nuclear import receptor protein family can reduce the formation of pathological TDP-43 aggregates. Using KPNB1 as a model, we found that its activity depends on the prion-like C-terminal region of TDP-43, which mediates the co-aggregation with phenylalanine and glycine-rich nucleoporins (FG-Nups) such as Nup62. KPNB1 is recruited into these co-aggregates where it acts as a molecular chaperone that reverses aberrant phase transition of Nup62 and TDP-43. These findings are supported by the discovery that Nup62 and KPNB1 are also sequestered into pathological TDP-43 aggregates in ALS/FTD postmortem CNS tissue, and by the identification of the fly ortholog of KPNB1 as a strong protective modifier in Drosophila models of TDP-43 proteinopathy. Our results show that KPNB1 can rescue all hallmarks of TDP-43 pathology, by restoring its solubility and nuclear localization, and reducing neurodegeneration in cellular and animal models of ALS/FTD.
Conclusion: Our findings suggest a novel NLS-independent mechanism where, analogous to its canonical role in dissolving the diffusion barrier formed by FG-Nups in the nuclear pore, KPNB1 is recruited into TDP-43/FG-Nup co-aggregates present in TDP-43 proteinopathies and therapeutically reverses their deleterious phase transition and mislocalization, mitigating neurodegeneration.
Keywords: Aggregation; Amyotrophic lateral sclerosis; Drosophila; Frontotemporal dementia; Importin; Nuclear pore; Nucleocytoplasmic transport; Prion-like domain; TDP-43.
© 2022. The Author(s).
Conflict of interest statement
The authors declare they have no financial competing interests. BK and WR are co-inventors on a patent relating to the content of the manuscript.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- U54NS123743/GF/NIH HHS/United States
- P01NS084974/GF/NIH HHS/United States
- R21 AG064940/AG/NIA NIH HHS/United States
- R01NS113943/GF/NIH HHS/United States
- R21 NS133676/NS/NINDS NIH HHS/United States
- R01AG059871/GF/NIH HHS/United States
- P30AG072931/RG/CSR NIH HHS/United States
- R21 NS096647/NS/NINDS NIH HHS/United States
- R01 NS105756/NS/NINDS NIH HHS/United States
- R33 NS110960/NS/NINDS NIH HHS/United States
- R01NS097542/GF/NIH HHS/United States
- R01NS105756/GF/NIH HHS/United States
- R01AG077771/GF/NIH HHS/United States
- R21AG064940/GF/NIH HHS/United States
- L30 AG048607/AG/NIA NIH HHS/United States
- R01NS117461/GF/NIH HHS/United States
- RF1AG068581/GF/NIH HHS/United States
- R35NS097273/GF/NIH HHS/United States
- RF1 AG068581/AG/NIA NIH HHS/United States
- R01NS127187/GF/NIH HHS/United States
- R21 NS100055/NS/NINDS NIH HHS/United States
- R21 NS098379/NS/NINDS NIH HHS/United States
- R01 AG059871/AG/NIA NIH HHS/United States
- T32 NS086749/NS/NINDS NIH HHS/United States
- R21NS096647/GF/NIH HHS/United States
- RF1 AG076122/AG/NIA NIH HHS/United States
- R33NS110960/GF/NIH HHS/United States
- R01 AG077771/AG/NIA NIH HHS/United States
- P30 AG072931/AG/NIA NIH HHS/United States
- P30AG062677/GF/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

