Profiling genome-wide recombination in Epstein Barr virus reveals type-specific patterns and associations with endemic-Burkitt lymphoma
- PMID: 36482473
- PMCID: PMC9733152
- DOI: 10.1186/s12985-022-01942-8
Profiling genome-wide recombination in Epstein Barr virus reveals type-specific patterns and associations with endemic-Burkitt lymphoma
Abstract
Background: Endemic Burkitt lymphoma (eBL) is potentiated through the interplay of Epstein Barr virus (EBV) and holoendemic Plasmodium falciparum malaria. To better understand EBV's biology and role in eBL, we characterized genome-wide recombination sites and patterns as a source of genetic diversity in EBV genomes in our well-defined population of eBL cases and controls from Western Kenya.
Methods: EBV genomes representing 54 eBL cases and 32 healthy children from the same geographic region in Western Kenya that we previously sequenced were analyzed. Whole-genome multiple sequence alignment, recombination analyses, and phylogenetic inference were made using multiple alignment with fast Fourier transform, recombination detection program 4, and molecular evolutionary genetics analysis.
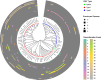
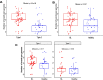
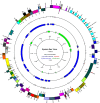
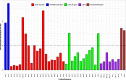
Results: We identified 28 different recombination events and 71 (82.6%) of the 86 EBV genomes analyzed contained evidence of one or more recombinant segments. Associated recombination breakpoints were found to occur in a total of 42 different genes, with only 7 (16.67%) being latent genes. Recombination events were major drivers of clustering within genome-wide phylogenetic trees. The occurrence of recombination segments was comparable between genomes from male and female participants and across age groups. More recombinant segments were found in EBV type 1 genomes (p = 6.4e - 06) and the genomes from the eBLs (p = 0.037). Two recombination events were enriched in the eBLs; event 47 (OR = 4.07, p = 0.038) and event 50 (OR = 14.24, p = 0.012).
Conclusions: EBV genomes have extensive evidence of recombination likely acquired progressively and cumulatively over time. Recombination patterns display a heterogeneous occurrence rate across the genome with enrichment in lytic genes. Overall, recombination appears to be a major evolutionary force impacting EBV diversity and genome structure with evidence of the association of specific recombinants with eBL.
Keywords: Endemic-Burkitt lymphoma; Epstein–Barr virus; Genome-wide recombination.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Epstein-Barr Virus Genomes Reveal Population Structure and Type 1 Association with Endemic Burkitt Lymphoma.J Virol. 2020 Aug 17;94(17):e02007-19. doi: 10.1128/JVI.02007-19. Print 2020 Aug 17. J Virol. 2020. PMID: 32581102 Free PMC article.
-
Epstein-Barr Virus in Burkitt Lymphoma in Africa Reveals a Limited Set of Whole Genome and LMP-1 Sequence Patterns: Analysis of Archival Datasets and Field Samples From Uganda, Tanzania, and Kenya.Front Oncol. 2022 Mar 7;12:812224. doi: 10.3389/fonc.2022.812224. eCollection 2022. Front Oncol. 2022. PMID: 35340265 Free PMC article.
-
Comprehensive Transcriptome and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV Type-Specific Differences.Mol Cancer Res. 2017 May;15(5):563-576. doi: 10.1158/1541-7786.MCR-16-0305. Mol Cancer Res. 2017. PMID: 28465297 Free PMC article.
-
Endemic Burkitt's lymphoma as a polymicrobial disease: new insights on the interaction between Plasmodium falciparum and Epstein-Barr virus.Semin Cancer Biol. 2009 Dec;19(6):411-20. doi: 10.1016/j.semcancer.2009.10.002. Epub 2009 Nov 6. Semin Cancer Biol. 2009. PMID: 19897039 Review.
-
The company malaria keeps: how co-infection with Epstein-Barr virus leads to endemic Burkitt lymphoma.Curr Opin Infect Dis. 2011 Oct;24(5):435-41. doi: 10.1097/QCO.0b013e328349ac4f. Curr Opin Infect Dis. 2011. PMID: 21885920 Free PMC article. Review.
References
-
- Moukassa D, Boumba AM, Ngatali CF, Ebatetou A, Mbon JBN, Ibara J-R. Virus-induced cancers in Africa: epidemiology and carcinogenesis mechanisms. Open J Pathol. 2018;08:1–14. doi: 10.4236/ojpathology.2018.81001. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

