Integrated transcriptome and metabolome profiling of Camellia reticulata reveal mechanisms of flower color differentiation
- PMID: 36482888
- PMCID: PMC9725097
- DOI: 10.3389/fgene.2022.1059717
Integrated transcriptome and metabolome profiling of Camellia reticulata reveal mechanisms of flower color differentiation
Abstract
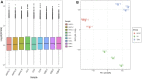
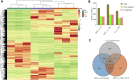
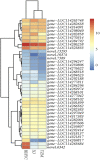
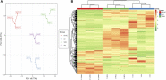
Camellia reticulata (Lindl.) is an important ornamental plant in China. Long-term natural or artificial selections have resulted in diverse phenotypes, especially for flower colors. Modulating flower colors can enhance the visual appeal and economic value in ornamental plants. In this study, we investigated the molecular mechanisms underlying flower color differentiation in C. reticulata. We performed a combined transcriptome and metabolome analysis of the petals of a popular variety C. reticulata (HHYC) (red), and its two cultivars "Xuejiao" (XJ) (pink) and "Tongzimian" (TZM) (white). Targeted metabolome profiling identified 310 flavonoid compounds of which 18 anthocyanins were differentially accumulated among the three samples with an accumulation pattern of HHYC > XJ > TZM. Likewise, transcriptome analysis showed that carotenoid and anthocyanin biosynthetic structural genes were mostly expressed in order of HHYC > XJ > TZM. Two genes (gene-LOC114287745765 and gene-LOC114289234) encoding for anthocyanidin 3-O-glucosyltransferase are predicted to be responsible for red coloration in HHYC and XJ. We also detected 42 MYB and 29 bHLH transcription factors as key regulators of anthocyanin-structural genes. Overall, this work showed that flavonoids, particularly anthocyanins contents are the major determinants of flower color differentiation among the 3 C. reticulata samples. In addition, the main regulatory and structural genes modulating anthocyanin contents in C. reticulata have been unveiled. Our results will help in the development of Camellia varieties with specific flower color and quality.
Keywords: Camellia reticulata; anthocyanins; carotenoids; flower; omics.
Copyright © 2022 Geng, Nie, Yang, Cai, Hu, Chen, Cheng, Wang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Coloration differences in three Camellia reticulata Lindl. cultivars: 'Tongzimian', 'Shizitou' and 'Damanao'.BMC Plant Biol. 2024 Jan 2;24(1):18. doi: 10.1186/s12870-023-04655-4. BMC Plant Biol. 2024. PMID: 38166751 Free PMC article.
-
Integrative metabolome and transcriptome analyses reveal the coloration mechanism in Camellia oleifera petals with different color.BMC Plant Biol. 2024 Jan 2;24(1):19. doi: 10.1186/s12870-023-04699-6. BMC Plant Biol. 2024. PMID: 38166635 Free PMC article.
-
Integrated metabolome and transcriptome analysis of the anthocyanin biosynthetic pathway in relation to color mutation in miniature roses.BMC Plant Biol. 2021 Jun 4;21(1):257. doi: 10.1186/s12870-021-03063-w. BMC Plant Biol. 2021. PMID: 34088264 Free PMC article.
-
Why Black Flowers? An Extreme Environment and Molecular Perspective of Black Color Accumulation in the Ornamental and Food Crops.Front Plant Sci. 2022 Apr 14;13:885176. doi: 10.3389/fpls.2022.885176. eCollection 2022. Front Plant Sci. 2022. PMID: 35498642 Free PMC article. Review.
-
Recent advances in flower color variation and patterning of Japanese morning glory and petunia.Breed Sci. 2018 Jan;68(1):128-138. doi: 10.1270/jsbbs.17107. Epub 2018 Mar 2. Breed Sci. 2018. PMID: 29681755 Free PMC article. Review.
Cited by
-
Metabolomics Analysis Reveals the Accumulation Patterns of Flavonoids and Volatile Compounds in Camellia oleifera Petals with Different Color.Molecules. 2023 Oct 24;28(21):7248. doi: 10.3390/molecules28217248. Molecules. 2023. PMID: 37959668 Free PMC article.
-
Coloration differences in three Camellia reticulata Lindl. cultivars: 'Tongzimian', 'Shizitou' and 'Damanao'.BMC Plant Biol. 2024 Jan 2;24(1):18. doi: 10.1186/s12870-023-04655-4. BMC Plant Biol. 2024. PMID: 38166751 Free PMC article.
-
Integration of Metabolomic and Transcriptomic Analyses Reveals the Molecular Mechanisms of Flower Color Formation in Prunus mume.Plants (Basel). 2024 Apr 11;13(8):1077. doi: 10.3390/plants13081077. Plants (Basel). 2024. PMID: 38674486 Free PMC article.
-
Characterization Variation of the Differential Coloring Substances in Rapeseed Petals with Different Colors Using UPLC-HESI-MS/MS.Molecules. 2023 Jul 26;28(15):5670. doi: 10.3390/molecules28155670. Molecules. 2023. PMID: 37570640 Free PMC article.
-
Integrated Transcriptomic and Metabolomic Analysis Reveals Nitrogen-Mediated Delay of Premature Leaf Senescence in Red Raspberry Leaves.Plants (Basel). 2025 Aug 2;14(15):2388. doi: 10.3390/plants14152388. Plants (Basel). 2025. PMID: 40805737 Free PMC article.
References
-
- Anders S. (2010). Analysing RNA-Seq data with the DESeq package. Mol. Biol. 43, 1–17.
-
- Beckles D. M., Roessner U. (2012). Plant metabolomics: Applications and opportunities for agricultural biotechnology. Plant Biotechnol. Agric., 67–81. 10.1016/B978-0-12-381466-1.00005-5 - DOI
-
- Ben-Simhon Z., Judeinstein S., Trainin T., Harel-Beja R., Bar-Yaakov I., Borochov-Neori H., et al. (2015). A “white” anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX; ANS) gene. PLoS One 10, e0142777. 10.1371/journal.pone.0142777 - DOI - PMC - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. 10.1111/J.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources

