AnnotaPipeline: An integrated tool to annotate eukaryotic proteins using multi-omics data
- PMID: 36482896
- PMCID: PMC9723129
- DOI: 10.3389/fgene.2022.1020100
AnnotaPipeline: An integrated tool to annotate eukaryotic proteins using multi-omics data
Abstract
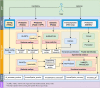
Assignment of gene function has been a crucial, laborious, and time-consuming step in genomics. Due to a variety of sequencing platforms that generates increasing amounts of data, manual annotation is no longer feasible. Thus, the need for an integrated, automated pipeline allowing the use of experimental data towards validation of in silico prediction of gene function is of utmost relevance. Here, we present a computational workflow named AnnotaPipeline that integrates distinct software and data types on a proteogenomic approach to annotate and validate predicted features in genomic sequences. Based on FASTA (i) nucleotide or (ii) protein sequences or (iii) structural annotation files (GFF3), users can input FASTQ RNA-seq data, MS/MS data from mzXML or similar formats, as the pipeline uses both transcriptomic and proteomic information to corroborate annotations and validate gene prediction, providing transcription and expression evidence for functional annotation. Reannotation of the available Arabidopsis thaliana, Caenorhabditis elegans, Candida albicans, Trypanosoma cruzi, and Trypanosoma rangeli genomes was performed using the AnnotaPipeline, resulting in a higher proportion of annotated proteins and a reduced proportion of hypothetical proteins when compared to the annotations publicly available for these organisms. AnnotaPipeline is a Unix-based pipeline developed using Python and is available at: https://github.com/bioinformatics-ufsc/AnnotaPipeline.
Keywords: functional annotation; genome annotation; hypothetical proteins; proteogenomics; workflow.
Copyright © 2022 Maia, Filho, Kawagoe, Teixeira Soratto, Moreira, Grisard and Wagner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources