Comparative glycoproteomics study on the surface of SKOV3 versus IOSE80 cell lines
- PMID: 36482940
- PMCID: PMC9723240
- DOI: 10.3389/fchem.2022.1010642
Comparative glycoproteomics study on the surface of SKOV3 versus IOSE80 cell lines
Abstract
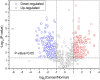
Objective: Site- and structure-specific quantitative N-glycoproteomics study of differential cell-surface N-glycosylation of ovarian cancer SKOV3 cells with the non-cancerous ovarian epithelial IOSE80 cells as the control. Methods: C18-RPLC-MS/MS (HCD with stepped normalized collision energies) was used to analyze the 1: 1 mixture of labeled intact N-glycopeptides from SKOV3 and IOSE80 cells, and the site- and structure-specific intact N-glycopeptide search engine GPSeeker was used to conduct qualitative and quantitative search on the obtained raw datasets. Results: With the control of the spectrum-level false discovery rate ≤1%, 13,822 glycopeptide spectral matches coming from 2,918 N-glycoproteins with comprehensive N-glycosite and N-glycan structure information were identified; 3,733 N-glycosites and 3,754 N-glycan sequence structures were confirmed by site-determining and structure-diagnostic fragment ions, respectively. With the control of no less than two observations among the three technical replicates, fold change ≥1.5, and p-value ≤ 0.05, 746 DEPGs in SKOV3 cells relative to IOSE80 cells were quantified, where 421 were upregulated and 325 downregulated. Conclusion: Differential cell-surface N-glycosylation of ovarian cancer SKOV3 cells were quantitatively analyzed by isotopic labeling and site- and structure-specific N-glycoproteomics. This discovery study provides putative N-glycoprotein biomarker candidates for future validation study using multiple reaction monitoring and biochemical methods.
Keywords: GPSeeker; differential N-glycosylation; quantitative N-glycoproteomics; site-specific; structure-specific.
Copyright © 2022 Zhou, Cai, Wu and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
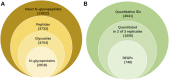

 = N-acetylglucosamine (Y),
= N-acetylglucosamine (Y), = mannose (M); au = arbitrary unit; IPMD = isotopic peak m/z deviation; IPAD = isotopic peak abundance deviation.
= mannose (M); au = arbitrary unit; IPMD = isotopic peak m/z deviation; IPAD = isotopic peak abundance deviation.
 =N-acetylglucosamine (Y);
=N-acetylglucosamine (Y); =mannose (M);
=mannose (M); = galactose (L);
= galactose (L); =sialic acid (S);
=sialic acid (S); = fucose (F); au = arbitrary unit; IPMD = isotopic peak m/z deviation; IPAD = isotopic peak abundance deviation.
= fucose (F); au = arbitrary unit; IPMD = isotopic peak m/z deviation; IPAD = isotopic peak abundance deviation.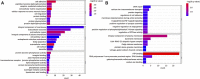

Similar articles
-
Site- and structure-specific quantitative N-glycoproteomics study of differential N-glycosylation in MCF-7 cancer cells.J Proteomics. 2020 Feb 10;212:103594. doi: 10.1016/j.jprot.2019.103594. Epub 2019 Nov 20. J Proteomics. 2020. PMID: 31759178
-
Site- and structure-specific characterization of the human urinary N-glycoproteome with site-determining and structure-diagnostic product ions.Rapid Commun Mass Spectrom. 2021 Jan 15;35(1):e8952. doi: 10.1002/rcm.8952. Rapid Commun Mass Spectrom. 2021. PMID: 32965048
-
GPSeeker Enables Quantitative Structural N-Glycoproteomics for Site- and Structure-Specific Characterization of Differentially Expressed N-Glycosylation in Hepatocellular Carcinoma.J Proteome Res. 2019 Jul 5;18(7):2885-2895. doi: 10.1021/acs.jproteome.9b00191. Epub 2019 Jun 3. J Proteome Res. 2019. PMID: 31117584
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Towards structure-focused glycoproteomics.Biochem Soc Trans. 2021 Feb 26;49(1):161-186. doi: 10.1042/BST20200222. Biochem Soc Trans. 2021. PMID: 33439247 Free PMC article. Review.
Cited by
-
Glycan diversity in ovarian cancer: Unraveling the immune interplay and therapeutic prospects.Semin Immunopathol. 2024 Oct 21;46(6):16. doi: 10.1007/s00281-024-01025-6. Semin Immunopathol. 2024. PMID: 39432076 Free PMC article. Review.
References
-
- Abbott K. L., Lim J. M., Wells L., Benigno B. B., McDonald J. F., Pierce M. (2010). Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 10 (3), 470–481. 10.1002/pmic.200900537 - DOI - PMC - PubMed
-
- Amano M., Yamaguchi M., Takegawa Y., Yamashita T., Terashima M., Furukawa J. i., et al. (2010). Threshold in stage-specific embryonic glycotypes uncovered by a full portrait of dynamic N-glycan expression during cell differentiation. Mol. Cell. Proteomics 9 (3), 523–537. 10.1074/mcp.m900559-mcp200 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

