IL-17A and TNF-α inhibitors induce multiple molecular changes in psoriasis
- PMID: 36483564
- PMCID: PMC9723344
- DOI: 10.3389/fimmu.2022.1015182
IL-17A and TNF-α inhibitors induce multiple molecular changes in psoriasis
Abstract
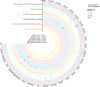
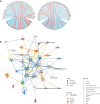
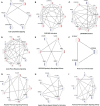
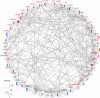
Adalimumab and secukinumab are commonly used for moderate to severe psoriasis vulgaris (PV). Although distinct individual responses to and impaired effectiveness of these biological agents occur occasionally, little is known about the underlying reasons. Here, we report a proteomic analysis of psoriatic lesions from patients treated with these drugs using data-independent acquisition mass spectrometry (DIA-MS). Thousands of differentially expressed proteins (DEPs) changed over 12 weeks of treatment. Network analysis showed that DEPs could interact and induce transformation in matrix components, metabolic regulation, and immune response. The results of parallel reaction monitoring (PRM) analysis suggested that S100s, STAT1, KRT2, TYMP, SOD2, HSP90AB1, TFRC, and COL5A1 were the most significantly changed proteins in both groups. There was a positive association between the Psoriasis Area and Severity Index (PASI) score and three proteins (TFRC, IMPDH2, KRT2). Our study findings suggest that inhibition of IL-17A and TNF-α can induce changes in multiple molecules in psoriatic lesions and have an overlapping influence on the immune response and process through direct or indirect effects.
Keywords: adalimumab; biological agent; data-independent acquisition mass spectrometry; ingenuity pathway analysis; parallel reaction monitoring; proteomics; psoriasis; secukinumab.
Copyright © 2022 Dong, Li, Xie, Hu, Huang, Jia, Tang, Liu, Shen and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

