The Caulobacter crescentus DciA promotes chromosome replication through topological loading of the DnaB replicative helicase at replication forks
- PMID: 36484102
- PMCID: PMC9825169
- DOI: 10.1093/nar/gkac1146
The Caulobacter crescentus DciA promotes chromosome replication through topological loading of the DnaB replicative helicase at replication forks
Abstract
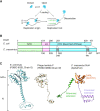
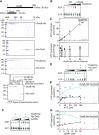
The replicative DNA helicase translocates on single-stranded DNA to drive replication forks during chromosome replication. In most bacteria the ubiquitous replicative helicase, DnaB, co-evolved with the accessory subunit DciA, but how they function remains incompletely understood. Here, using the model bacterium Caulobacter crescentus, we demonstrate that DciA plays a prominent role in DNA replication fork maintenance. Cell cycle analyses using a synchronized Caulobacter cell population showed that cells devoid of DciA exhibit a severe delay in fork progression. Biochemical characterization revealed that the DnaB helicase in its default state forms a hexamer that inhibits self-loading onto single-stranded DNA. We found that upon binding to DciA, the DnaB hexamer undergoes conformational changes required for encircling single-stranded DNA, thereby establishing the replication fork. Further investigation of the functional structure of DciA revealed that the C-terminus of DciA includes conserved leucine residues responsible for DnaB binding and is essential for DciA in vivo functions. We propose that DciA stimulates loading of DnaB onto single strands through topological isomerization of the DnaB structure, thereby ensuring fork progression. Given that the DnaB-DciA modules are widespread among eubacterial species, our findings suggest that a common mechanism underlies chromosome replication.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Katayama T., Ozaki S., Keyamura K., Fujimitsu K.. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010; 8:163–170. - PubMed

