The impact of modifier genes on cone-rod dystrophy heterogeneity: An explorative familial pilot study and a hypothesis on neurotransmission impairment
- PMID: 36490268
- PMCID: PMC9733859
- DOI: 10.1371/journal.pone.0278857
The impact of modifier genes on cone-rod dystrophy heterogeneity: An explorative familial pilot study and a hypothesis on neurotransmission impairment
Abstract
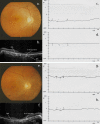
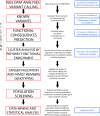
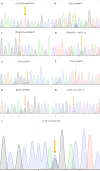
Cone-rod dystrophies (CORDs) are a heterogeneous group of inherited retinopathies (IRDs) with more than 30 already known disease-causing genes. Uncertain phenotypes and extended range of intra- and interfamilial heterogenicity make still difficult to determine a precise genotype-phenotype correlation. Here, we used a next-generation sequencing approach to study a Sicilian family with a suspected form of CORD. Affected family members underwent ophthalmological examinations and a proband, blind from 50 years, underwent whole genome and exome sequencing. Variant analysis was enriched by pathway analysis and relevant variants were, then, investigated in other family members and in 100 healthy controls from Messina. CORD diagnosis with an intricate pattern of symptoms was confirmed by ophthalmological examinations. A total of about 50,000 variants were identified in both proband's genome and exome. All affected family members presented specific genotypes mainly determined by mutated GUCY2D gene, and different phenotypical traits, mainly related to focus and color perception. Thus, we looked for possible modifier genes. According to relationship with GUCY2D, predicted functional effects, eye localization, and ocular disease affinity, only 9 variants, carried by 6 genes (CACNG8, PAX2, RXRG, CCDC175, PDE4DIP and LTF), survived the filtering. These genes encode key proteins involved in cone development and survival, and retina neurotransmission. Among analyzed variants, CACNG8c.*6819A>T and the new CCDC175 c.76C>T showed extremely low frequency in the control group, suggesting a key role on disease phenotypes. Such discovery could enforce the role of modifier genes into CORD onset/progression, contributing to improve diagnostic test towards a better personalized medicine.
Copyright: © 2022 Donato et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Comprehensive Genotyping and Phenotyping Analysis of GUCY2D-Associated Rod- and Cone-Dominated Dystrophies.Am J Ophthalmol. 2023 Oct;254:87-103. doi: 10.1016/j.ajo.2023.05.015. Epub 2023 Jun 15. Am J Ophthalmol. 2023. PMID: 37327959
-
Identification of a novel RPGR mutation associated with X-linked cone-rod dystrophy in a Chinese family.BMC Ophthalmol. 2021 Nov 20;21(1):401. doi: 10.1186/s12886-021-02166-0. BMC Ophthalmol. 2021. PMID: 34800980 Free PMC article.
-
Variants at codon 838 in the GUCY2D gene result in different phenotypes of cone rod dystrophy.Ophthalmic Genet. 2020 Dec;41(6):548-555. doi: 10.1080/13816810.2020.1807026. Epub 2020 Aug 18. Ophthalmic Genet. 2020. PMID: 32811265
-
Whole exome sequencing reveals GUCY2D as a major gene associated with cone and cone-rod dystrophy in Israel.Invest Ophthalmol Vis Sci. 2014 Dec 16;56(1):420-30. doi: 10.1167/iovs.14-15647. Invest Ophthalmol Vis Sci. 2014. PMID: 25515582 Free PMC article.
-
Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: mutation spectrum and new genotype-phenotype correlation.Orphanet J Rare Dis. 2015 Jun 24;10:85. doi: 10.1186/s13023-015-0300-3. Orphanet J Rare Dis. 2015. PMID: 26103963 Free PMC article.
Cited by
-
Transcriptional patterns of human retinal pigment epithelial cells under protracted high glucose.Mol Biol Rep. 2024 Apr 4;51(1):477. doi: 10.1007/s11033-024-09479-5. Mol Biol Rep. 2024. PMID: 38573426
-
Investigating G-quadruplex structures in RPGR gene: Implications for understanding X-linked retinal degeneration.Heliyon. 2024 Apr 18;10(8):e29828. doi: 10.1016/j.heliyon.2024.e29828. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38699732 Free PMC article.
-
Inhibition of mTOR differently modulates planar and subepithelial fibrogenesis in human conjunctival fibroblasts.Graefes Arch Clin Exp Ophthalmol. 2025 Jan;263(1):33-46. doi: 10.1007/s00417-024-06481-2. Epub 2024 Jul 23. Graefes Arch Clin Exp Ophthalmol. 2025. PMID: 39042147
-
Transcriptome Analysis of Retinal and Choroidal Pathologies in Aged BALB/c Mice Following Systemic Neonatal Murine Cytomegalovirus Infection.Int J Mol Sci. 2023 Feb 21;24(5):4322. doi: 10.3390/ijms24054322. Int J Mol Sci. 2023. PMID: 36901754 Free PMC article.
-
Causal Associations of Glaucoma and Age-Related Macular Degeneration with Cataract: A Bidirectional Two-Sample Mendelian Randomisation Study.Genes (Basel). 2024 Mar 26;15(4):413. doi: 10.3390/genes15040413. Genes (Basel). 2024. PMID: 38674349 Free PMC article.
References
-
- Gill JS, Georgiou M, Kalitzeos A, Moore AT, Michaelides M. Progressive cone and cone-rod dystrophies: clinical features, molecular genetics and prospects for therapy. The British journal of ophthalmology. 2019. Epub 20190124. doi: 10.1136/bjophthalmol-2018-313278 ; PubMed Central PMCID: PMC6709772. - DOI - PMC - PubMed
-
- Liew G, Michaelides M, Bunce C. A comparison of the causes of blindness certifications in England and Wales in working age adults (16–64 years), 1999–2000 with 2009–2010. BMJ Open. 2014;4(2):e004015. Epub 20140212. doi: 10.1136/bmjopen-2013-004015 ; PubMed Central PMCID: PMC3927710. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

