NARF is a hypoxia-induced coactivator for OCT4-mediated breast cancer stem cell specification
- PMID: 36490339
- PMCID: PMC9733926
- DOI: 10.1126/sciadv.abo5000
NARF is a hypoxia-induced coactivator for OCT4-mediated breast cancer stem cell specification
Abstract
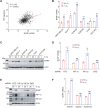
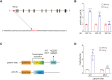
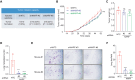
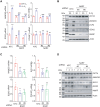
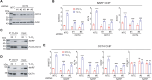
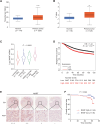
Hypoxia is a key characteristic of the breast cancer microenvironment that promotes expression of the transcriptional activator hypoxia-inducible factor 1 (HIF-1) and is associated with poor patient outcome. HIF-1 increases the expression or activity of stem cell pluripotency factors, which control breast cancer stem cell (BCSC) specification and are required for cancer metastasis. Here, we identify nuclear prelamin A recognition factor (NARF) as a hypoxia-inducible, HIF-1 target gene in human breast cancer cells. NARF functions as an essential coactivator by recruiting the histone demethylase KDM6A to OCT4 bound to genes encoding the pluripotency factors NANOG, KLF4, and SOX2, leading to demethylation of histone H3 trimethylated at lysine-27 (H3K27me3), thereby increasing the expression of NANOG, KLF4, and SOX2, which, together with OCT4, mediate BCSC specification. Knockdown of NARF significantly decreased the BCSC population in vitro and markedly impaired tumor initiation capacity and lung metastasis in orthotopic mouse models.
Figures




References
-
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray,Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71,209–249 (2021). - PubMed
-
- G. P. Gupta, J. Massagué,Cancer metastasis: Building a framework. Cell 127,679–695 (2006). - PubMed
-
- P. S. Steeg,Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 12,895–904 (2006). - PubMed
-
- R. Bos, H. Zhong, C. F. Hanrahan, F. C. Mommers, G. L. Semenza, H. M. Pinedo, M. D. Abeloff, J. W. Simons, P. J. van Diest, E. van der Wall,Levels of hypoxia-inducible factor-1α during breast carcinogenesis. J. Natl. Cancer Inst. 93,309–314 (2001). - PubMed
-
- A. L. Harris,Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2,38–47 (2002). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

