Leveraging an established neighbourhood-level, open access wastewater monitoring network to address public health priorities: a population-based study
- PMID: 36493788
- PMCID: PMC9725778
- DOI: 10.1016/S2666-5247(22)00289-0
Leveraging an established neighbourhood-level, open access wastewater monitoring network to address public health priorities: a population-based study
Abstract
Background: Before the COVID-19 pandemic, the US opioid epidemic triggered a collaborative municipal and academic effort in Tempe, Arizona, which resulted in the world's first open access dashboard featuring neighbourhood-level trends informed by wastewater-based epidemiology (WBE). This study aimed to showcase how wastewater monitoring, once established and accepted by a community, could readily be adapted to respond to newly emerging public health priorities.
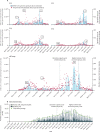
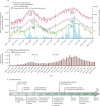
Methods: In this population-based study in Greater Tempe, Arizona, an existing opioid monitoring WBE network was modified to track SARS-CoV-2 transmission through the analysis of 11 contiguous wastewater catchments. Flow-weighted and time-weighted 24 h composite samples of untreated wastewater were collected at each sampling location within the wastewater collection system for 3 days each week (Tuesday, Thursday, and Saturday) from April 1, 2020, to March 31, 2021 (Area 7 and Tempe St Luke's Hospital were added in July, 2020). Reverse transcription quantitative PCR targeting the E gene of SARS-CoV-2 isolated from the wastewater samples was used to determine the number of genome copies in each catchment. Newly detected clinical cases of COVID-19 by zip code within the City of Tempe, Arizona were reported daily by the Arizona Department of Health Services from May 23, 2020. Maricopa County-level new positive cases, COVID-19-related hospitalisations, deaths, and long-term care facility deaths per day are publicly available and were collected from the Maricopa County Epidemic Curve Dashboard. Viral loads of SARS-CoV-2 (genome copies per day) measured in wastewater from each catchment were aggregated at the zip code level and city level and compared with the clinically reported data using root mean square error to investigate early warning capability of WBE.
Findings: Between April 1, 2020, and March 31, 2021, 1556 wastewater samples were analysed. Most locations showed two waves in viral levels peaking in June, 2020, and December, 2020-January, 2021. An additional wave of viral load was seen in catchments close to Arizona State University (Areas 6 and 7) at the beginning of the fall (autumn) semester in late August, 2020. Additionally, an early infection hotspot was detected in the Town of Guadalupe, Arizona, starting the week of May 4, 2020, that was successfully mitigated through targeted interventions. A shift in early warning potential of WBE was seen, from a leading (mean of 8·5 days [SD 2·1], June, 2020) to a lagging (-2·0 days [1·4], January, 2021) indicator compared with newly reported clinical cases.
Interpretation: Lessons learned from leveraging an existing neighbourhood-level WBE reporting dashboard include: (1) community buy-in is key, (2) public data sharing is effective, and (3) sub-ZIP-code (postal code) data can help to pinpoint populations at risk, track intervention success in real time, and reveal the effect of local clinical testing capacity on WBE's early warning capability. This successful demonstration of transitioning WBE efforts from opioids to COVID-19 encourages an expansion of WBE to tackle newly emerging and re-emerging threats (eg, mpox and polio).
Funding: National Institutes of Health's RADx-rad initiative, National Science Foundation, Virginia G Piper Charitable Trust, J M Kaplan Fund, and The Flinn Foundation.
Copyright © 2023 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests EMD is a managing member of AquaVitas, a company working in the field of wastewater-based epidemiology. RUH is a managing member of AquaVitas and founder of the Arizona State University non-profit project OneWaterOneHealth operating in the same intellectual space. All other authors declare no competing interests.
Figures

References
-
- Gracia-Lor E, Castiglioni S, Bade R, et al. Measuring biomarkers in wastewater as a new source of epidemiological information: current state and future perspectives. Environ Int. 2017;99:131–150. - PubMed
-
- Choi PM, Tscharke BJ, Donner E, et al. Wastewater-based epidemiology biomarkers: past, present and future. Trends Analyt Chem. 2018;105:453–469.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

