Machine Learning Modeling of Protein-intrinsic Features Predicts Tractability of Targeted Protein Degradation
- PMID: 36494034
- PMCID: PMC10025769
- DOI: 10.1016/j.gpb.2022.11.008
Machine Learning Modeling of Protein-intrinsic Features Predicts Tractability of Targeted Protein Degradation
Abstract
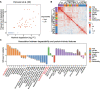
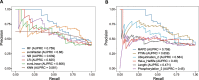
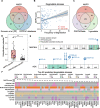
Targeted protein degradation (TPD) has rapidly emerged as a therapeutic modality to eliminate previously undruggable proteins by repurposing the cell's endogenous protein degradation machinery. However, the susceptibility of proteins for targeting by TPD approaches, termed "degradability", is largely unknown. Here, we developed a machine learning model, model-free analysis of protein degradability (MAPD), to predict degradability from features intrinsic to protein targets. MAPD shows accurate performance in predicting kinases that are degradable by TPD compounds [with an area under the precision-recall curve (AUPRC) of 0.759 and an area under the receiver operating characteristic curve (AUROC) of 0.775] and is likely generalizable to independent non-kinase proteins. We found five features with statistical significance to achieve optimal prediction, with ubiquitination potential being the most predictive. By structural modeling, we found that E2-accessible ubiquitination sites, but not lysine residues in general, are particularly associated with kinase degradability. Finally, we extended MAPD predictions to the entire proteome to find 964 disease-causing proteins (including proteins encoded by 278 cancer genes) that may be tractable to TPD drug development.
Keywords: Degradability; Machine learning; Protein-intrinsic feature; Targeted protein degradation; Ubiquitination.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
X. Shirley Liu is a cofounder, board member, and CEO of GV20 Therapeutics. Eric S. Fischer is a founder, member of the scientific advisory board (SAB), and equity holder of Civetta Therapeutics, Lighthorse Therapeutics, Proximity Therapeutics, and Neomorph Inc (board of directors), SAB member and equity holder in Avilar Therapeutics and Photys Therapeutics, and a consultant to Astellas, Sanofi, Novartis, Deerfield, and EcoR1 capital. The Fischer laboratory receives or has received research funding from Novartis, Deerfield, Ajax, Interline, and Astellas. Katherine A. Donovan is a consultant to Kronos Bio and Neomorph Inc. All the other authors declared no competing interests.
Figures



References
-
- Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
-
- Baumeister W., Walz J., Zühl F., Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
-
- Burslem G.M., Crews C.M. Small-molecule modulation of protein homeostasis. Chem Rev. 2017;117:11269–11301. - PubMed
-
- Liu J., Farmer J.D., Jr, Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

