Defective proteostasis in induced pluripotent stem cell models of frontotemporal lobar degeneration
- PMID: 36494352
- PMCID: PMC9734180
- DOI: 10.1038/s41398-022-02274-5
Defective proteostasis in induced pluripotent stem cell models of frontotemporal lobar degeneration
Abstract
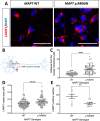
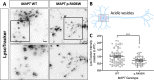
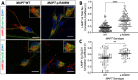
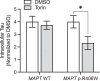
Impaired proteostasis is associated with normal aging and is accelerated in neurodegeneration. This impairment may lead to the accumulation of protein, which can be toxic to cells and tissue. In a subset of frontotemporal lobar degeneration with tau pathology (FTLD-tau) cases, pathogenic mutations in the microtubule-associated protein tau (MAPT) gene are sufficient to cause tau accumulation and neurodegeneration. However, the pathogenic events triggered by the expression of the mutant tau protein remain poorly understood. Here, we show that molecular networks associated with lysosomal biogenesis and autophagic function are disrupted in brains from FTLD-tau patients carrying a MAPT p.R406W mutation. We then used human induced pluripotent stem cell (iPSC)-derived neurons and 3D cerebral organoids from patients carrying the MAPT p.R406W mutation and CRISPR/Cas9, corrected controls to evaluate proteostasis. MAPT p.R406W was sufficient to induce morphological and functional deficits in the lysosomal pathway in iPSC-neurons. These phenotypes were reversed upon correction of the mutant allele with CRISPR/Cas9. Treatment with mTOR inhibitors led to tau degradation specifically in MAPT p.R406W neurons. Together, our findings suggest that MAPT p.R406W is sufficient to cause impaired lysosomal function, which may contribute to disease pathogenesis and serve as a cellular phenotype for drug screening.
© 2022. The Author(s).
Conflict of interest statement
ST is president of StemCultures, scientific co-founder of Luxa Biotech, and has served on the scientific advisory boards of Sana Biotechnology and Blue Rock Therapeutics and as a consultant to Merck. KH is an Eisai-sponsored visiting researcher at Washington University and has received salary from Eisai. The remaining authors declare no competing interests.
Figures




References
-
- Goedert M, Jakes R. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta. 2005;1739:240–50. - PubMed
-
- Reed LA, Grabowski TJ, Schmidt ML, Morris JC, Goate A, Solodkin A, et al. Autosomal dominant dementia with widespread neurofibrillary tangles. Ann Neurol. 1997;42:564–72. - PubMed
-
- van Swieten JC, Stevens M, Rosso SM, Rizzu P, Joosse M, de Koning I, et al. Phenotypic variation in hereditary frontotemporal dementia with tau mutations. Ann Neurol. 1999;46:617–26. - PubMed
-
- Lindquist SG, Holm IE, Schwartz M, Law I, Stokholm J, Batbayli M, et al. Alzheimer disease-like clinical phenotype in a family with FTDP-17 caused by a MAPT R406W mutation. Eur J Neurol. 2008;15:377–85. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

