Whole genome sequencing reveals new links between spa t172/CC59 methicillin-resistant Staphylococcus aureus cases in low-endemicity region of Southwest Finland, 2007‒2016
- PMID: 36494398
- PMCID: PMC9734107
- DOI: 10.1038/s41598-022-25556-w
Whole genome sequencing reveals new links between spa t172/CC59 methicillin-resistant Staphylococcus aureus cases in low-endemicity region of Southwest Finland, 2007‒2016
Abstract
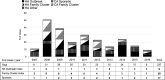
Methicillin-resistant Staphylococcus aureus (MRSA) rates have remained relatively low in Finland. In Southwest Finland, however, annual MRSA incidence increased from 12 to 25/100,000 between 2007 and 2016 with spa t172 strain causing one fourth (237/983) of all cases. This provoked us to study the molecular epidemiology of t172-MRSA, aiming to better understand the transmission of this strain type. We combined epidemiological data and whole genome sequencing (WGS) of a set of 64 (27%, 64/237) t172-MRSA isolates covering 10 years. Isolates represented sporadic and index cases of all identified healthcare-associated outbreaks (HAOs) and family clusters (FCs). Among the included 62 isolates, core-genome MLST analysis revealed eight genomic clusters comprising 24 (38.7%) isolates and 38 (61.3%) non-clustered isolates. Cluster 1 comprised ten and the remaining seven clusters two isolates each, respectively. Two epidemiologically distinct HAOs were linked in cluster 1. FCs were involved in all clusters. All strains were associated with epidemic clonal complex CC59. We were able to confirm the spread of several successful t172-MRSA subclones in regional healthcare and the community. WGS complemented routine surveillance by revealing undetected links between t172-MRSA cases. Targeted, WGS-based typing could enhance MRSA surveillance without the need for routine WGS diagnostics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Cassini A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA in 2019 (EARS-Net)–Annual Epidemiological Report 2019. (2020).
-
- Finnish Institute for Health and Welfare. Infectious Diseases in Finland 2021 (in Finnish). (2022).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

