Variable-selection ANOVA Simultaneous Component Analysis (VASCA)
- PMID: 36495189
- PMCID: PMC9825241
- DOI: 10.1093/bioinformatics/btac795
Variable-selection ANOVA Simultaneous Component Analysis (VASCA)
Abstract
Motivation: ANOVA Simultaneous Component Analysis (ASCA) is a popular method for the analysis of multivariate data yielded by designed experiments. Meaningful associations between factors/interactions of the experimental design and measured variables in the dataset are typically identified via significance testing, with permutation tests being the standard go-to choice. However, in settings with large numbers of variables, like omics (genomics, transcriptomics, proteomics and metabolomics) experiments, the 'holistic' testing approach of ASCA (all variables considered) often overlooks statistically significant effects encoded by only a few variables (biomarkers).
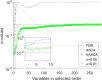
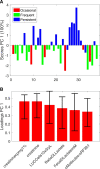
Results: We hereby propose Variable-selection ASCA (VASCA), a method that generalizes ASCA through variable selection, augmenting its statistical power without inflating the Type-I error risk. The method is evaluated with simulations and with a real dataset from a multi-omic clinical experiment. We show that VASCA is more powerful than both ASCA and the widely adopted false discovery rate controlling procedure; the latter is used as a benchmark for variable selection based on multiple significance testing. We further illustrate the usefulness of VASCA for exploratory data analysis in comparison to the popular partial least squares discriminant analysis method and its sparse counterpart.
Availability and implementation: The code for VASCA is available in the MEDA Toolbox at https://github.com/josecamachop/MEDA-Toolbox (release v1.3). The simulation results and motivating example can be reproduced using the repository at https://github.com/josecamachop/VASCA/tree/v1.0.0 (DOI 10.5281/zenodo.7410623).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures


References
-
- Anderson M., Braak C.T. (2003) Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Simul., 73, 85–113.
-
- Barker M., Rayens W. (2003) Partial least squares for discrimination. J. Chemometr., 17, 166–173.
-
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological), 57, 289–300.
-
- Benjamini Y., Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann. Stat., 29, 1165–1188.
-
- Bevilacqua M. et al. (2013) Application of near infrared (NIR) spectroscopy coupled to chemometrics for dried egg-pasta characterization and egg content quantification. Food Chem., 140, 726–734. - PubMed

