Limits to transcriptional silencing in Saccharomyces cerevisiae
- PMID: 36495285
- PMCID: PMC9910407
- DOI: 10.1093/genetics/iyac180
Limits to transcriptional silencing in Saccharomyces cerevisiae
Abstract
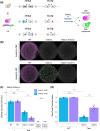
Mating-type switching in the budding yeast Saccharomyces cerevisiae relies on the Sir protein complex to silence HML and HMR, the two loci containing copies of the alleles of the mating type locus, MAT. Sir-based transcriptional silencing has been considered locus-specific, but the recent discovery of rare and transient escapes from silencing at HMLα2 with a sensitive assay called to question if these events extend to the whole locus. Adapting the same assay, we measured that transient silencing failures at HML were more frequent for the α2 gene than α1, similarly to their expression level in unsilenced cells. By coupling a mating assay, at HML we found that one of the two genes at that locus can be transiently expressed while the other gene is maintained silent. Thus, transient silencing loss can be a property of the gene rather than the locus. Cells lacking the SIR1 gene experience epigenetic bistability at HML and HMR. Our previous result led us to ask if HML could allow for two independent epigenetic states within the locus in a sir1Δ mutant. A simple construct using a double fluorescent reporter at HMLα1 and HMLα2 ruled out this possibility. Each HML locus displayed a single epigenetic state. We revisited the question of the correlation between the states of two HML loci in diploid cells, and showed they were independent. Finally, we determined the relative strength of gene repression achieved by Sir-based silencing with that achieved by the a1-α2 repressor.
Keywords: bistability; epigenetics; heterochromatin; mating-type regulation; recombination enhancer; sirtuins; transcriptional silencing.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest: The authors declare no conflict of interest.
Figures




References
-
- Benaglia T, Chauveau D, Hunter DR, Young DS. Mixtools: an R package for analyzing mixture models. J Stat Softw. 2009;32(6):1–29. doi:10.18637/jss.v032.i06. - DOI
-
- Bergh D. Sample size and chi-squared test of fit—a comparison between a random sample approach and a chi-square value adjustment method using Swedish adolescent data. In: Zhang Q, Yang H, editors. Pacific Rim Objective Measurement Symposium (PROMS) 2014 Conference Proceedings. Berlin (Germany: ): Springer; 2015. p. 197–211.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

