m6A Profile Dynamics Indicates Regulation of Oyster Development by m6A-RNA Epitranscriptomes
- PMID: 36496129
- PMCID: PMC10787124
- DOI: 10.1016/j.gpb.2022.12.002
m6A Profile Dynamics Indicates Regulation of Oyster Development by m6A-RNA Epitranscriptomes
Erratum in
-
Correction to: m6A Profile Dynamics Indicates Regulation of Oyster Development by m6A-RNA Epitranscriptomes.Genomics Proteomics Bioinformatics. 2024 Jul 3;22(2):qzae021. doi: 10.1093/gpbjnl/qzae021. Genomics Proteomics Bioinformatics. 2024. PMID: 38981582 Free PMC article. No abstract available.
Abstract
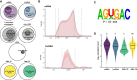
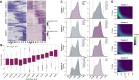
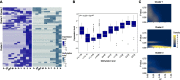
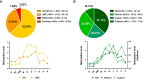
The N6-methylation of RNA adenosines (N6-methyladenosine, m6A) is an important regulator of gene expression with critical implications in vertebrate and insect development. However, the developmental significance of epitranscriptomes in lophotrochozoan organisms remains unknown. Using methylated RNA immunoprecipitation sequencing (MeRIP-seq), we generated transcriptome-wide m6A-RNA methylomes covering the entire development of the oyster from oocytes to juveniles. Oyster RNA classes display specific m6A signatures, with messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) exhibiting distinct profiles and being highly methylated compared to transposable element (TE) transcripts. Epitranscriptomes are dynamic and correspond to the chronological steps of development (cleavage, gastrulation, organogenesis, and metamorphosis), with minimal mRNA and lncRNA methylation at the morula stage followed by a global increase. mRNA m6A levels are correlated with transcript levels, and shifts in methylation profiles correspond to expression kinetics. Differentially methylated transcripts cluster according to embryo-larval stages and bear the corresponding developmental functions (cell division, signal transduction, morphogenesis, and cell differentiation). The m6A level of TE transcripts is also regulated and peaks during the gastrulation. We demonstrate that m6A-RNA methylomes are dynamic and associated with gene expression regulation during oyster development. The putative epitranscriptome implication in the cleavage, maternal-to-zygotic transition, and cell differentiation in a lophotrochozoan model brings new insights into the control and evolution of developmental processes.
Keywords: Embryo; Evo-devo; Metamorphosis; RNA methylation; Transcription.
Copyright © 2022 Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have declared no competing interests.
Figures


Similar articles
-
Dynamics of DNA methylomes underlie oyster development.PLoS Genet. 2017 Jun 8;13(6):e1006807. doi: 10.1371/journal.pgen.1006807. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28594821 Free PMC article.
-
N6-Methyladenosine Methylation Analysis of Long Noncoding RNAs and mRNAs in IPEC-J2 Cells Treated With Clostridium perfringens beta2 Toxin.Front Immunol. 2021 Nov 22;12:769204. doi: 10.3389/fimmu.2021.769204. eCollection 2021. Front Immunol. 2021. PMID: 34880865 Free PMC article.
-
A functional m6 A-RNA methylation pathway in the oyster Crassostrea gigas assumes epitranscriptomic regulation of lophotrochozoan development.FEBS J. 2021 Mar;288(5):1696-1711. doi: 10.1111/febs.15500. Epub 2020 Sep 1. FEBS J. 2021. PMID: 32743927
-
N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases.FEBS J. 2016 May;283(9):1607-30. doi: 10.1111/febs.13614. Epub 2015 Dec 31. FEBS J. 2016. PMID: 26645578 Review.
-
N1-methyladenosine methylome in messenger RNA and non-coding RNA.Curr Opin Chem Biol. 2018 Aug;45:179-186. doi: 10.1016/j.cbpa.2018.06.017. Epub 2018 Jul 9. Curr Opin Chem Biol. 2018. PMID: 30007213 Review.
Cited by
-
Core genes of biomineralization and cis-regulatory long non-coding RNA regulate shell growth in bivalves.J Adv Res. 2024 Oct;64:117-129. doi: 10.1016/j.jare.2023.11.024. Epub 2023 Nov 22. J Adv Res. 2024. PMID: 37995944 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

