Mitochondrial dysfunction induces ALK5-SMAD2-mediated hypovascularization and arteriovenous malformations in mouse retinas
- PMID: 36496409
- PMCID: PMC9741628
- DOI: 10.1038/s41467-022-35262-w
Mitochondrial dysfunction induces ALK5-SMAD2-mediated hypovascularization and arteriovenous malformations in mouse retinas
Abstract
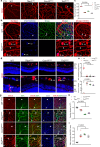
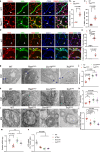
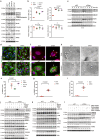
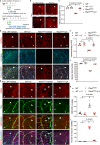
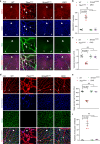
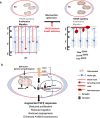
Although mitochondrial activity is critical for angiogenesis, its mechanism is not entirely clear. Here we show that mice with endothelial deficiency of any one of the three nuclear genes encoding for mitochondrial proteins, transcriptional factor (TFAM), respiratory complex IV component (COX10), or redox protein thioredoxin 2 (TRX2), exhibit retarded retinal vessel growth and arteriovenous malformations (AVM). Single-cell RNA-seq analyses indicate that retinal ECs from the three mutant mice have increased TGFβ signaling and altered gene expressions associated with vascular maturation and extracellular matrix, correlating with vascular malformation and increased basement membrane thickening in microvesels of mutant retinas. Mechanistic studies suggest that mitochondrial dysfunction from Tfam, Cox10, or Trx2 depletion induces a mitochondrial localization and MAPKs-mediated phosphorylation of SMAD2, leading to enhanced ALK5-SMAD2 signaling. Importantly, pharmacological blockade of ALK5 signaling or genetic deficiency of SMAD2 prevented retinal vessel growth retardation and AVM in all three mutant mice. Our studies uncover a novel mechanism whereby mitochondrial dysfunction via the ALK5-SMAD2 signaling induces retinal vascular malformations, and have therapeutic values for the alleviation of angiogenesis-associated human retinal diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Potente, M., Gerhardt, H. & Carmeliet, P. Basic and therapeutic aspects of angiogenesis. Cell146, 873–887 (2011). - PubMed
-
- Adams, R. H. & Alitalo, K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol.8, 464–478 (2007). - PubMed
-
- Ferrara, N. & Kerbel, R. S. Angiogenesis as a therapeutic target. Nature438, 967–974 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

