Daily Rhythms in the IGF-1 System in the Liver of Goldfish and Their Synchronization to Light/Dark Cycle and Feeding Time
- PMID: 36496892
- PMCID: PMC9739714
- DOI: 10.3390/ani12233371
Daily Rhythms in the IGF-1 System in the Liver of Goldfish and Their Synchronization to Light/Dark Cycle and Feeding Time
Abstract
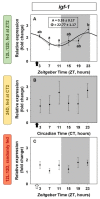
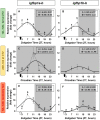
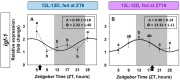
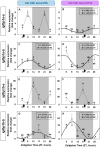
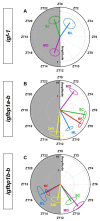
The relevance of the insulin-like growth factor-1 (IGF-1) system in several physiological processes is well-known in vertebrates, although little information about their temporal organization is available. This work aims to investigate the possible rhythmicity of the different components of the IGF-1 system (igf-1, the igf1ra and igf1rb receptors and the paralogs of its binding proteins IGFBP1 and IGFBP2) in the liver of goldfish. In addition, we also study the influence of two environmental cues, the light/dark cycle and feeding time, as zeitgebers. The hepatic igf-1 expression showed a significant daily rhythm with the acrophase prior to feeding time, which seems to be strongly dependent on both zeitgebers. Only igfbp1a-b and igfbp1b-b paralogs exhibited a robust daily rhythm of expression in the liver that persists in fish held under constant darkness or randomly fed. The hepatic expression of the two receptor subtypes did not show daily rhythms in any of the experimental conditions. Altogether these results point to the igf-1, igfbp1a-b, and igfbp1b-b as clock-controlled genes, supporting their role as putative rhythmic outputs of the hepatic oscillator, and highlight the relevance of mealtime as an external cue for the 24-h rhythmic expression of the IGF-1 system in fish.
Keywords: biological clock; chronobiology; circadian; fish; gene expression; insulin-like binding proteins; insulin-like growth factor 1; insulin-like growth factor receptors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Triantaphyllopoulos K.A., Cartas D., Miliou H. Factors influencing GH and IGF-I gene expression on growth in teleost fish: How can aquaculture industry benefit? Rev. Aquac. 2019;12:1637–1662. doi: 10.1111/raq.12402. - DOI
-
- Shimizu M. Insulin-Like Growth Factor-1. In: Ando H., Ukena K., Nagata S., editors. Handbook of Hormones. Comparative Endocrinology for Basic and Clinical Research. Academic Press; London, UK: 2021. pp. 285–296.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

