Anatomical Development of the Cerebellothalamic Tract in Embryonic Mice
- PMID: 36497060
- PMCID: PMC9738252
- DOI: 10.3390/cells11233800
Anatomical Development of the Cerebellothalamic Tract in Embryonic Mice
Abstract
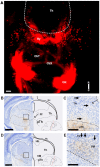
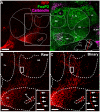
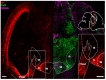
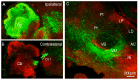
The main connection from cerebellum to cerebrum is formed by cerebellar nuclei axons that synapse in the thalamus. Apart from its role in coordinating sensorimotor integration in the adult brain, the cerebello-thalamic tract (CbT) has also been implicated in developmental disorders, such as autism spectrum disorders. Although the development of the cerebellum, thalamus and cerebral cortex have been studied, there is no detailed description of the ontogeny of the mammalian CbT. Here we investigated the development of the CbT at embryonic stages using transgenic Ntsr1-Cre/Ai14 mice and in utero electroporation of wild type mice. Wide-field, confocal and 3D light-sheet microscopy of immunohistochemical stainings showed that CbT fibers arrive in the prethalamus between E14.5 and E15.5, but only invade the thalamus after E16.5. We quantified the spread of CbT fibers throughout the various thalamic nuclei and found that at E17.5 and E18.5 the ventrolateral, ventromedial and parafascicular nuclei, but also the mediodorsal and posterior complex, become increasingly innervated. Several CbT fiber varicosities express vesicular glutamate transporter type 2 at E18.5, indicating cerebello-thalamic synapses. Our results provide the first quantitative data on the developing murine CbT, which provides guidance for future investigations of the impact that cerebellum has on thalamo-cortical networks during development.
Keywords: cerebello-thalamic tract; cerebellum; embryonic development; mouse; thalamus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. - DOI - PMC - PubMed
-
- Jacobowitz D.M., Abbott L.C. Chemoarchitectonic Atlas of the Developing Mouse Brain. CRC Press; Boca Raton, FL, USA: 1998.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

