The Revelation of Continuously Organized, Co-Overexpressed Protein-Coding Genes with Roles in Cellular Communications in Breast Cancer
- PMID: 36497066
- PMCID: PMC9741223
- DOI: 10.3390/cells11233806
The Revelation of Continuously Organized, Co-Overexpressed Protein-Coding Genes with Roles in Cellular Communications in Breast Cancer
Abstract
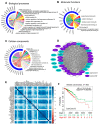
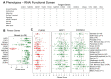
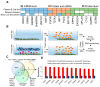
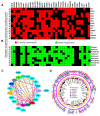
Many human cancers, including breast cancer, are polygenic and involve the co-dysregulation of multiple regulatory molecules and pathways. Though the overexpression of genes and amplified chromosomal regions have been closely linked in breast cancer, the notion of the co-upregulation of genes at a single locus remains poorly described. Here, we describe the co-overexpression of 34 continuously organized protein-coding genes with diverse functions at 8q.24.3(143437655-144326919) in breast and other cancer types, the CanCord34 genes. In total, 10 out of 34 genes have not been reported to be overexpressed in breast cancer. Interestingly, the overexpression of CanCord34 genes is not necessarily associated with genomic amplification and is independent of hormonal or HER2 status in breast cancer. CanCord34 genes exhibit diverse known and predicted functions, including enzymatic activities, cell viability, multipotency, cancer stem cells, and secretory activities, including extracellular vesicles. The co-overexpression of 33 of the CanCord34 genes in a multivariant analysis was correlated with poor survival among patients with breast cancer. The analysis of the genome-wide RNAi functional screening, cell dependency fitness, and breast cancer stem cell databases indicated that three diverse overexpressed CanCord34 genes, including a component of spliceosome PUF60, a component of exosome complex EXOSC4, and a ribosomal biogenesis factor BOP1, shared roles in cell viability, cell fitness, and stem cell phenotypes. In addition, 17 of the CanCord34 genes were found in the microvesicles (MVs) secreted from the mesenchymal stem cells that were primed with MDA-MB-231 breast cancer cells. Since these MVs were important in the chemoresistance and dedifferentiation of breast cancer cells into cancer stem cells, these findings highlight the significance of the CanCord34 genes in cellular communications. In brief, the persistent co-overexpression of CanCord34 genes with diverse functions can lead to the dysregulation of complementary functions in breast cancer. In brief, the present study provides new insights into the polygenic nature of breast cancer and opens new research avenues for basic, preclinical, and therapeutic studies in human cancer.
Keywords: breast cancer; cancer progression; computational biology; coregulation; emerging pathways; polygenic nature; shared mechanisms of gene expression; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448.BMC Cancer. 2016 Feb 25;16:160. doi: 10.1186/s12885-016-2108-5. BMC Cancer. 2016. PMID: 26917489 Free PMC article.
-
Quantitative proteomic analysis of HER2 normal and overexpressing MCF-7 breast cancer cells revealed proteomic changes accompanied with HER2 gene amplification.J Proteomics. 2013 Oct 8;91:200-9. doi: 10.1016/j.jprot.2013.06.034. Epub 2013 Jul 11. J Proteomics. 2013. PMID: 23851309
-
Novel signaling molecules implicated in tumor-associated fatty acid synthase-dependent breast cancer cell proliferation and survival: Role of exogenous dietary fatty acids, p53-p21WAF1/CIP1, ERK1/2 MAPK, p27KIP1, BRCA1, and NF-kappaB.Int J Oncol. 2004 Mar;24(3):591-608. Int J Oncol. 2004. PMID: 14767544
-
Basal-Type Breast Cancer Stem Cells Over-Express Chromosomal Passenger Complex Proteins.Cells. 2020 Mar 13;9(3):709. doi: 10.3390/cells9030709. Cells. 2020. PMID: 32183150 Free PMC article.
-
Tissue factor mediates microvesicles shedding from MDA-MB-231 breast cancer cells.Biochem Biophys Res Commun. 2018 Jul 7;502(1):137-144. doi: 10.1016/j.bbrc.2018.05.136. Epub 2018 May 24. Biochem Biophys Res Commun. 2018. PMID: 29787758
Cited by
-
Hyperactivation of p21-Activated Kinases in Human Cancer and Therapeutic Sensitivity.Biomedicines. 2023 Feb 5;11(2):462. doi: 10.3390/biomedicines11020462. Biomedicines. 2023. PMID: 36830998 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

