Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer's Disease
- PMID: 36497090
- PMCID: PMC9741225
- DOI: 10.3390/cells11233830
Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer's Disease
Abstract
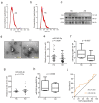
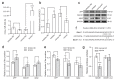
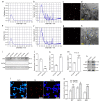
Alzheimer's disease (AD) is a common neurodegenerative disorder with progressive cognitive impairment in the elderly. Beta-amyloid (Aβ) formation and its accumulation in the brain constitute one of the pathological hallmarks of AD. Until now, how to modulate Aβ formation in hippocampal neurons remains a big challenge. Herein, we investigated whether the exosomal transfer of microRNA (miR) relates to amyloid pathology in the recipient neuron cells. We isolated circulating small extracellular vesicles (sEVs) from AD patients and healthy controls, determined the miR-342-5p level in the sEVs by RT-PCR, and evaluated its diagnostic performance in AD. Then, we took advantage of biomolecular assays to estimate the role of miR-342-5p in modulating the amyloid pathway, including amyloid precursor protein (APP), beta-site APP cleaving enzyme 1 (BACE1), and Aβ42. Furthermore, we subjected HT22 cells to the sEVs from the hippocampal tissues of transgenic APP mice (Exo-APP) or C57BL/6 littermates (Exo-CTL), and the Exo-APP enriched with miR-342-5p mimics or the control to assess the effect of the sEVs' delivery of miR-342-5p on Aβ formation. We observed a lower level of miR-342-5p in the circulating sEVs from AD patients compared with healthy controls. MiR-342-5p participated in Aβ formation by modulating BACE1 expression, specifically binding its 3'-untranslated region (UTR) sequence. Exo-APP distinctly promoted Aβ42 formation in the recipient cells compared to Exo-CTL. Intriguingly, miR-342-5p enrichment in Exo-APP ameliorated amyloid pathology in the recipient cells. Our study indicated that miR-342-5p was dysregulated in human circulating sEVs from AD patients; sEV transfer of miR-342-5p ameliorates Aβ formation by modulating BACE1 expression. These findings highlight the promising potential of exosomal miRNAs in AD clinical therapy.
Keywords: Alzheimer’s disease; BACE1; amyloid-beta; extracellular vesicle; miRNA; neurons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
MicroRNA-339-5p down-regulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects.J Biol Chem. 2014 Feb 21;289(8):5184-98. doi: 10.1074/jbc.M113.518241. Epub 2013 Dec 18. J Biol Chem. 2014. PMID: 24352696 Free PMC article.
-
MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer's disease.Int J Mol Med. 2014 Jul;34(1):160-6. doi: 10.3892/ijmm.2014.1780. Epub 2014 May 13. Int J Mol Med. 2014. PMID: 24827165
-
miR-98-5p Acts as a Target for Alzheimer's Disease by Regulating Aβ Production Through Modulating SNX6 Expression.J Mol Neurosci. 2016 Dec;60(4):413-420. doi: 10.1007/s12031-016-0815-7. Epub 2016 Aug 19. J Mol Neurosci. 2016. PMID: 27541017
-
Alzheimer's disease.Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review.
-
The beta-secretase, BACE: a prime drug target for Alzheimer's disease.J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
Cited by
-
Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer's Disease.Int J Mol Sci. 2023 Nov 13;24(22):16259. doi: 10.3390/ijms242216259. Int J Mol Sci. 2023. PMID: 38003448 Free PMC article. Review.
-
A new perspective on Alzheimer's disease: microRNAs and circular RNAs.Front Genet. 2023 Sep 15;14:1231486. doi: 10.3389/fgene.2023.1231486. eCollection 2023. Front Genet. 2023. PMID: 37790702 Free PMC article. Review.
-
Advancements in extracellular vesicle therapy for neurodegenerative diseases.Explor Neuroprotective Ther. 2025;5:10.37349/ent.2025.1004104. doi: 10.37349/ent.2025.1004104. Epub 2025 May 6. Explor Neuroprotective Ther. 2025. PMID: 40894255 Free PMC article.
-
Bioengineered MSC-derived exosomes in skin wound repair and regeneration.Front Cell Dev Biol. 2023 Feb 27;11:1029671. doi: 10.3389/fcell.2023.1029671. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36923255 Free PMC article. Review.
-
Exosomes: A Cellular Communication Medium That Has Multiple Effects On Brain Diseases.Mol Neurobiol. 2024 Sep;61(9):6864-6892. doi: 10.1007/s12035-024-03957-4. Epub 2024 Feb 14. Mol Neurobiol. 2024. PMID: 38356095 Review.
References
-
- Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., Loeloff R., et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

