Genomewide m6A Mapping Uncovers Dynamic Changes in the m6A Epitranscriptome of Cisplatin-Treated Apoptotic HeLa Cells
- PMID: 36497162
- PMCID: PMC9738315
- DOI: 10.3390/cells11233905
Genomewide m6A Mapping Uncovers Dynamic Changes in the m6A Epitranscriptome of Cisplatin-Treated Apoptotic HeLa Cells
Abstract
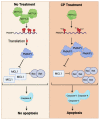
Cisplatin (CP), which is a conventional cancer chemotherapeutic drug, induces apoptosis by modulating a diverse array of gene regulatory mechanisms. However, cisplatin-mediated changes in the m6A methylome are unknown. We employed an m6A miCLIP-seq approach to investigate the effect of m6A methylation marks under cisplatin-mediated apoptotic conditions on HeLa cells. Our high-resolution approach revealed numerous m6A marks on 972 target mRNAs with an enrichment on 132 apoptotic mRNAs. We tracked the fate of differentially methylated candidate mRNAs under METTL3 knockdown and cisplatin treatment conditions. Polysome profile analyses revealed perturbations in the translational efficiency of PMAIP1 and PHLDA1 transcripts. Congruently, PMAIP1 amounts were dependent on METTL3. Additionally, cisplatin-mediated apoptosis was sensitized by METTL3 knockdown. These results suggest that apoptotic pathways are modulated by m6A methylation events and that the METTL3-PMAIP1 axis modulates cisplatin-mediated apoptosis in HeLa cells.
Keywords: HeLa; apoptosis; cisplatin; epitranscriptomics; m6A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Epitranscriptomics m6A analyses reveal distinct m6A marks under tumor necrosis factor α (TNF-α)-induced apoptotic conditions in HeLa cells.J Cell Physiol. 2024 Apr;239(4):e31176. doi: 10.1002/jcp.31176. Epub 2024 Jan 5. J Cell Physiol. 2024. PMID: 38179601
-
METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP.Cell Biol Int. 2020 Dec;44(12):2524-2531. doi: 10.1002/cbin.11459. Epub 2020 Sep 11. Cell Biol Int. 2020. PMID: 32869897
-
Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis.Cells. 2022 Sep 24;11(19):2980. doi: 10.3390/cells11192980. Cells. 2022. PMID: 36230944 Free PMC article.
-
N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases.FEBS J. 2016 May;283(9):1607-30. doi: 10.1111/febs.13614. Epub 2015 Dec 31. FEBS J. 2016. PMID: 26645578 Review.
-
The METTL3-m6A Epitranscriptome: Dynamic Regulator of Epithelial Development, Differentiation, and Cancer.Genes (Basel). 2021 Jun 30;12(7):1019. doi: 10.3390/genes12071019. Genes (Basel). 2021. PMID: 34209046 Free PMC article. Review.
Cited by
-
Expression patterns of m6A RNA methylation regulators under apoptotic conditions in various human cancer cell lines.Turk J Biol. 2023 Dec 14;48(1):24-34. doi: 10.55730/1300-0152.2679. eCollection 2024. Turk J Biol. 2023. PMID: 38665783 Free PMC article.
-
The METTL3/TRAP1 axis as a key regulator of 5-fluorouracil chemosensitivity in colorectal cancer.Mol Cell Biochem. 2025 Mar;480(3):1865-1889. doi: 10.1007/s11010-024-05116-8. Epub 2024 Sep 17. Mol Cell Biochem. 2025. PMID: 39287889 Free PMC article.
-
HN1 Functions in Protein Synthesis Regulation via mTOR-RPS6 Axis and Maintains Nucleolar Integrity.Cell Prolif. 2025 Jun;58(6):e13805. doi: 10.1111/cpr.13805. Epub 2025 Jan 13. Cell Prolif. 2025. PMID: 39805577 Free PMC article.
-
YTHDF1 Promotes Bladder Cancer Cell Proliferation via the METTL3/YTHDF1-RPN2-PI3K/AKT/mTOR Axis.Int J Mol Sci. 2023 Apr 7;24(8):6905. doi: 10.3390/ijms24086905. Int J Mol Sci. 2023. PMID: 37108067 Free PMC article.
-
Uncovering the Epitranscriptome: A Review on mRNA Modifications and Emerging Frontiers.Genes (Basel). 2025 Aug 12;16(8):951. doi: 10.3390/genes16080951. Genes (Basel). 2025. PMID: 40870000 Free PMC article. Review.
References
-
- Ming X., Groehler I.A., Michaelson-Richie E.D., Villalta P.W., Campbell C., Tretyakova N.Y. Mass Spectrometry Based Proteomics Study of Cisplatin-Induced DNA–Protein Cross-Linking in Human Fibrosarcoma (HT1080) Cells. Chem. Res. Toxicol. 2017;30:980–995. doi: 10.1021/acs.chemrestox.6b00389. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

