A Signaling Crosstalk Links SNAIL to the 37/67 kDa Laminin-1 Receptor Ribosomal Protein SA and Regulates the Acquisition of a Cancer Stem Cell Molecular Signature in U87 Glioblastoma Neurospheres
- PMID: 36497426
- PMCID: PMC9738384
- DOI: 10.3390/cancers14235944
A Signaling Crosstalk Links SNAIL to the 37/67 kDa Laminin-1 Receptor Ribosomal Protein SA and Regulates the Acquisition of a Cancer Stem Cell Molecular Signature in U87 Glioblastoma Neurospheres
Erratum in
-
Correction: Gresseau et al. A Signaling Crosstalk Links SNAIL to the 37/67 kDa Laminin-1 Receptor Ribosomal Protein SA and Regulates the Acquisition of a Cancer Stem Cell Molecular Signature in U87 Glioblastoma Neurospheres. Cancers 2022, 14, 5944.Cancers (Basel). 2024 Mar 6;16(5):1065. doi: 10.3390/cancers16051065. Cancers (Basel). 2024. PMID: 38473439 Free PMC article.
Abstract
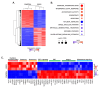
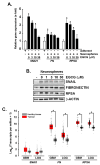
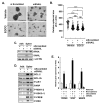
Background: Three-dimensional in vitro neurospheres cultures recapitulate stemness features associated with poor clinical outcome in glioblastoma patients. They are commonly used to address brain cancer stem cell (CSC) signal transducing biology that regulates spheroids formation and stemness phenotype, and to assess the in vitro pharmacological impact of chemotherapeutic drugs. Objective: Here, we addressed the role of a new signaling axis involved in the regulation of in vitro spheroids formation and assessed the chemopreventive ability of diet-derived epigallocatechin gallate (EGCG) to impact the processes that govern the acquisition of spheroids CSC stemness traits. Methods: Neurospheres were generated from adherent human U87 glioblastoma cancer cell cultures under conditions that recapitulate stemness features. Total RNA and protein lysates were isolated for gene expression by RT-qPCR and protein expression by immunoblot. Transcriptomic analysis was performed through RNA-Seq. Results: Compared to their parental adherent cells, tumorspheres expressed increased levels of the CSC markers NANOG, SOX2, PROM1 (CD133), as well as of the epithelial-to-mesenchymal transition (EMT) markers Fibronectin, SNAI1, and 37/67 kDa laminin-1 receptor ribosomal protein SA (RPSA). Increased PROM1, SOX2, Fibronectin, and RPSA transcripts level were also observed in clinical grade IV glioblastoma tissues compared to normal tissue. EGCG treatment reduced dose-dependently tumorspheres size and inhibited the transcriptional regulation of those genes. An apoptotic signature was also found in spheroids with increased signal transducing events involving GSK3α/β, RSK, and CREB. These were repressed upon RPSA gene silencing and partially by SNAI1 silencing. Conclusion: This work highlights a signaling axis linking RPSA upstream of SNAIL in neurospheres genesis and supports the chemopreventive impact that diet-derived EGCG may exert on the acquisition of CSC traits.
Keywords: EGCG; EMT; RPSA; SNAIL; cancer stem cells; glioblastoma; laminin receptor; spheroids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Tamimi A.F., Juweid M. Epidemiology and Outcome of Glioblastoma. In: De Vleeschouwer S., editor. Glioblastoma. Codon Publications; Brisbane, Australia: 2017. Chapter 8. - PubMed
-
- Alves A.L.V., Gomes I.N.F., Carloni A.C., Rosa M.N., da Silva L.S., Evangelista A.F., Reis R.M., Silva V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021;12:206. doi: 10.1186/s13287-021-02231-x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

