ACAP1 Deficiency Predicts Inferior Immunotherapy Response in Solid Tumors
- PMID: 36497434
- PMCID: PMC9740925
- DOI: 10.3390/cancers14235951
ACAP1 Deficiency Predicts Inferior Immunotherapy Response in Solid Tumors
Abstract
Background: ACAP1 plays a key role in endocytic recycling, which is essential for the normal function of lymphocytes. However, the expression and function of ACAP1 in lymphocytes have rarely been studied.
Methods: Large-scale genomic data, including multiple bulk RNA-sequencing datasets, single-cell sequencing datasets, and immunotherapy cohorts, were exploited to comprehensively characterize ACAP1 expression, regulation, and function. Gene set enrichment analysis (GSEA) was used to uncover the pathways associated with ACAP1 expression. Eight algorithms, including TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, xCELL, MCPCOUNTER, EPIC, and TIDE, were applied to estimate the infiltrating level of immune cells. Western blotting, qPCR, and ChIP-PCR were used to validate the findings from bioinformatic analyses. A T-cell co-culture killing assay was used to investigate the function of ACAP1 in lymphocytes.
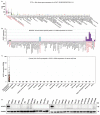
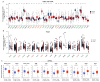
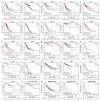
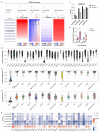
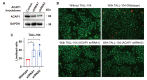
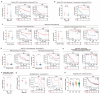
Results: ACAP1 was highly expressed in immune-related tissues and cells and minimally in other tissues. Moreover, single-cell sequencing analysis in tumor samples revealed that ACAP1 is expressed primarily in tumor-infiltrating lymphocytes (TILs), including T, B, and NK cells. ACAP1 expression is negatively regulated by promoter DNA methylation, with its promoter hypo-methylated in immune cells but hyper-methylated in other cells. Furthermore, SPI1 binds to the ACAP1 promoter and positively regulates its expression in immune cells. ACAP1 levels positively correlate with the infiltrating levels of TILs, especially CD8+ T cells, across a broad range of solid cancer types. ACAP1 deficiency is associated with poor prognosis and immunotherapeutic response in multiple cancer types treated with checkpoint blockade therapy (ICT). Functionally, the depletion of ACAP1 by RNA interference significantly impairs the T cell-mediated killing of tumor cells.
Conclusions: Our study demonstrates that ACAP1 is essential for the normal function of TILs, and its deficiency indicates an immunologically "cold" status of tumors that are resistant to ICT.
Keywords: ACAP1; CD8+ T cells; cancer immunotherapy; immune checkpoint blockade therapy (ICT); tumor-infiltrating lymphocytes (TILs).
Conflict of interest statement
Authors declare no conflict of interests.
Figures


Similar articles
-
Integrated analysis and validation reveal ACAP1 as a novel prognostic biomarker associated with tumor immunity in lung adenocarcinoma.Comput Struct Biotechnol J. 2022 Aug 13;20:4390-4401. doi: 10.1016/j.csbj.2022.08.026. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051873 Free PMC article.
-
Association of Germline Variants in Natural Killer Cells With Tumor Immune Microenvironment Subtypes, Tumor-Infiltrating Lymphocytes, Immunotherapy Response, Clinical Outcomes, and Cancer Risk.JAMA Netw Open. 2019 Sep 4;2(9):e199292. doi: 10.1001/jamanetworkopen.2019.9292. JAMA Netw Open. 2019. PMID: 31483464 Free PMC article.
-
Expression of ACAP1 Is Associated with Tumor Immune Infiltration and Clinical Outcome of Ovarian Cancer.DNA Cell Biol. 2020 Sep;39(9):1545-1557. doi: 10.1089/dna.2020.5596. Epub 2020 May 22. DNA Cell Biol. 2020. PMID: 32456571
-
The role of tumor-infiltrating lymphocytes in cholangiocarcinoma.J Exp Clin Cancer Res. 2022 Apr 7;41(1):127. doi: 10.1186/s13046-022-02340-2. J Exp Clin Cancer Res. 2022. PMID: 35392957 Free PMC article. Review.
-
[Function of tumor infiltrating lymphocytes in solid tumors - a review].Sheng Wu Gong Cheng Xue Bao. 2019 Dec 25;35(12):2308-2325. doi: 10.13345/j.cjb.190300. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31880138 Review. Chinese.
Cited by
-
Targeting epigenetic regulators as a promising avenue to overcome cancer therapy resistance.Signal Transduct Target Ther. 2025 Jul 18;10(1):219. doi: 10.1038/s41392-025-02266-z. Signal Transduct Target Ther. 2025. PMID: 40675967 Free PMC article. Review.
-
IGF2BP3 prevent HMGB1 mRNA decay in bladder cancer and development.Cell Mol Biol Lett. 2024 Mar 19;29(1):39. doi: 10.1186/s11658-024-00545-1. Cell Mol Biol Lett. 2024. PMID: 38504159 Free PMC article.
-
Recent Advancements in Research on DNA Methylation and Testicular Germ Cell Tumors: Unveiling the Intricate Relationship.Biomedicines. 2024 May 8;12(5):1041. doi: 10.3390/biomedicines12051041. Biomedicines. 2024. PMID: 38791003 Free PMC article. Review.
-
Identification and validation of a genetic risk signature associated with prognosis in clear-cell renal cell carcinoma patients.Medicine (Baltimore). 2023 Aug 4;102(31):e34582. doi: 10.1097/MD.0000000000034582. Medicine (Baltimore). 2023. PMID: 37543772 Free PMC article.
-
Clinical Proteomics Reveals Vulnerabilities in Noninvasive Breast Ductal Carcinoma and Drives Personalized Treatment Strategies.Cancer Res Commun. 2025 Jan 1;5(1):138-149. doi: 10.1158/2767-9764.CRC-24-0287. Cancer Res Commun. 2025. PMID: 39723668 Free PMC article.
References
-
- McDermott D.F., Huseni M.A., Atkins M.B., Motzer R.J., Rini B.I., Escudier B., Fong L., Joseph R.W., Pal S.K., Reeves J.A., et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018;24:749–757. doi: 10.1038/s41591-018-0053-3. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

