PKCβ Facilitates Leukemogenesis in Chronic Lymphocytic Leukaemia by Promoting Constitutive BCR-Mediated Signalling
- PMID: 36497487
- PMCID: PMC9735720
- DOI: 10.3390/cancers14236006
PKCβ Facilitates Leukemogenesis in Chronic Lymphocytic Leukaemia by Promoting Constitutive BCR-Mediated Signalling
Abstract
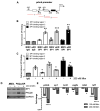
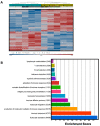
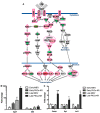
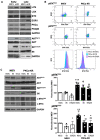
B cell antigen receptor (BCR) signalling competence is critical for the pathogenesis of chronic lymphocytic leukaemia (CLL). Defining key proteins that facilitate these networks aid in the identification of targets for therapeutic exploitation. We previously demonstrated that reduced PKCα function in mouse hematopoietic stem/progenitor cells (HPSCs) resulted in PKCβII upregulation and generation of a poor-prognostic CLL-like disease. Here, prkcb knockdown in HSPCs leads to reduced survival of PKCα-KR-expressing CLL-like cells, concurrent with reduced expression of the leukemic markers CD5 and CD23. SP1 promotes elevated expression of prkcb in PKCα-KR expressing cells enabling leukemogenesis. Global gene analysis revealed an upregulation of genes associated with B cell activation in PKCα-KR expressing cells, coincident with upregulation of PKCβII: supported by activation of key signalling hubs proximal to the BCR and elevated proliferation. Ibrutinib (BTK inhibitor) or enzastaurin (PKCβII inhibitor) treatment of PKCα-KR expressing cells and primary CLL cells showed similar patterns of Akt/mTOR pathway inhibition, supporting the role for PKCβII in maintaining proliferative signals in our CLL mouse model. Ibrutinib or enzastaurin treatment also reduced PKCα-KR-CLL cell migration towards CXCL12. Overall, we demonstrate that PKCβ expression facilitates leukemogenesis and identify that BCR-mediated signalling is a key driver of CLL development in the PKCα-KR model.
Keywords: BCR signalling; CLL; PKCβ; SP1; leukemogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

