Peak Plasma Levels of mtDNA Serve as a Predictive Biomarker for COVID-19 in-Hospital Mortality
- PMID: 36498735
- PMCID: PMC9740249
- DOI: 10.3390/jcm11237161
Peak Plasma Levels of mtDNA Serve as a Predictive Biomarker for COVID-19 in-Hospital Mortality
Abstract
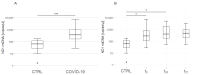
Several predictive biomarkers for coronavirus disease (COVID-19)-associated mortality in critically ill patients have been described. Although mitochondrial DNA (mtDNA) is elevated in patients with COVID-19, the association with coagulation function and its predictive power for mortality is unclear. Accordingly, this study investigates the predictive power of mtDNA for in-hospital mortality in critically ill patients with COVID-19, and whether combining it with thromboelastographic parameters can increase its predictive performance. This prospective explorative study included 29 patients with COVID-19 and 29 healthy matched controls. mtDNA encoding for NADH dehydrogenase 1 (ND1) was quantified using a quantitative polymerase chain reaction analysis, while coagulation function was evaluated using thromboelastometry and impedance aggregometry. Receiver operating characteristic (ROC) curves were used for the prediction of in-hospital mortality. Within the first 24 h, the plasma levels of mtDNA peaked significantly (controls: 65 (28-119) copies/µL; patients: 281 (110-805) at t0, 403 (168-1937) at t24, and 467 (188-952) copies/µL at t72; controls vs. patients: p = 0.02 at t0, p = 0.03 at t24, and p = 0.44 at t72). The mtDNA levels at t24 showed an excellent predictive performance for in-hospital mortality (area under the ROC curve: 0.90 (0.75-0.90)), which could not be improved by the combination with thromboelastometric or aggregometric parameters. Critically ill patients with COVID-19 present an early increase in the plasma levels of ND1 mtDNA, lasting over 24 h. They also show impairments in platelet function and fibrinolysis, as well as hypercoagulability, but these do not correlate with the plasma levels of fibrinogen. The peak plasma levels of mtDNA can be used as a predictive biomarker for in-hospital mortality; however, the combination with coagulation parameters does not improve the predictive validity.
Keywords: SARS-CoV2; biomarker; immunothrombosis; intensive care unit; nucleic acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- World Health Organisation Coronavirus Disease (COVID-19) Weekly Epidemiological Update. [(accessed on 2 September 2022)]. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio....
-
- Bonaventura A., Vecchié A., Dagna L., Martinod K., Dixon D.L., van Tassell B.W., Dentali F., Montecucco F., Massberg S., Levi M., et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 2021;21:319–329. doi: 10.1038/s41577-021-00536-9. - DOI - PMC - PubMed
-
- Evans P.C., Rainger G.E., Mason J.C., Guzik T.J., Osto E., Stamataki Z., Neil D., Hoefer I.E., Fragiadaki M., Waltenberger J., et al. Endothelial dysfunction in COVID-19: A position paper of the ESC Working Group for Atherosclerosis and Vascular Biology, and the ESC Council of Basic Cardiovascular Science. Cardiovasc. Res. 2020;116:2177–2184. doi: 10.1093/cvr/cvaa230. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

