Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas
- PMID: 36498835
- PMCID: PMC9735924
- DOI: 10.3390/ijms232314507
Transcriptional Profiling Provides New Insights into Organ Culture-Induced Changes in Human Donor Corneas
Abstract
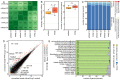
Corneal transplantation is one of the most common forms of tissue transplantation worldwide. Donor corneal tissue used in transplantation is provided by eye banks, which store the tissue in culture medium after procurement. To date, the effects of cell culture on human corneal tissue have not been fully elucidated. Using the 3' RNA sequencing method for massive analysis of cDNA ends (MACE), we show that cultivation of corneal tissue leads to significant changes in a variety of molecular processes in human corneal tissue that go well beyond aspects of previously known culture effects. Functionally grouped network analysis revealed nine major groups of biological processes that were affected by corneal organ culture, among them keratinization, hypoxia, and angiogenesis, with genes from each group being affected by culture time. A cell type deconvolution analysis revealed significant modulations of the corneal immune cell profile in a time dependent manner. The results suggest that current culture conditions should be further refined and that prolonged cultivation may be detrimental. Recently, we showed that MACE enables transcriptional profiling of formalin-fixed and paraffin-embedded (FFPE) conjunctival tissue with high accuracy even after more than 10 years of storage. Here we demonstrate that MACE provides comparable results for native and FFPE corneal tissue, confirming that the technology is suitable for transcriptome analysis of a wide range of archived diseased corneal samples stored in histological archives. Finally, our data underscore the feasibility of bioinformatics cell-type enrichment analysis in bulk RNA-seq data to profile immune cell composition in fixed and archived corneal tissue samples, for which RNA-seq analysis of individual cells is often not possible.
Keywords: RNA sequencing; cornea; corneal graft; corneal transplantation; eye bank; formalin-fixed paraffin-embedded; organ culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Wolf J., Boneva S., Schlecht A., Lapp T., Auw-Haedrich C., Lagrèze W., Agostini H., Reinhard T., Schlunck G., Lange C. The Human Eye Transcriptome Atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics. 2022;114:110286. doi: 10.1016/j.ygeno.2022.110286. - DOI - PubMed
-
- Collin J., Queen R., Zerti D., Bojic S., Dorgau B., Moyse N., Molina M.M., Yang C., Dey S., Reynolds G., et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 2021;21:279–298. doi: 10.1016/j.jtos.2021.03.010. - DOI - PMC - PubMed
-
- Boneva S., Schlecht A., Böhringer D., Mittelviefhaus H., Reinhard T., Agostini H., Auw-Haedrich C., Schlunck G., Wolf J., Lange C. 3′ MACE RNA-sequencing allows for transcriptome profiling in human tissue samples after long-term storage. Lab. Investig. 2020;100:1345–1355. doi: 10.1038/s41374-020-0446-z. - DOI - PMC - PubMed
-
- EEBA Minimum Medical Standards (MMS). Published Online 1 February 2020. [(accessed on 31 May 2022)]. Available online: https://www.eeba.eu/files/pdf/EEBA%20Minimum%20Medical%20Standards%20Rev....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

