19S Proteasome Subunits as Oncogenes and Prognostic Biomarkers in FLT3-Mutated Acute Myeloid Leukemia (AML)
- PMID: 36498916
- PMCID: PMC9740165
- DOI: 10.3390/ijms232314586
19S Proteasome Subunits as Oncogenes and Prognostic Biomarkers in FLT3-Mutated Acute Myeloid Leukemia (AML)
Abstract
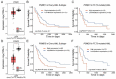
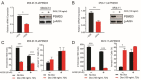
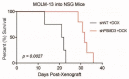
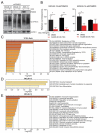
26S proteasome non-ATPase subunits 1 (PSMD1) and 3 (PSMD3) were recently identified as prognostic biomarkers and potential therapeutic targets in chronic myeloid leukemia (CML) and multiple solid tumors. In the present study, we analyzed the expression of 19S proteasome subunits in acute myeloid leukemia (AML) patients with mutations in the FMS-like tyrosine kinase 3 (FLT3) gene and assessed their impact on overall survival (OS). High levels of PSMD3 but not PSMD1 expression correlated with a worse OS in FLT3-mutated AML. Consistent with an oncogenic role for PSMD3 in AML, shRNA-mediated PSMD3 knockdown impaired colony formation of FLT3+ AML cell lines, which correlated with increased OS in xenograft models. While PSMD3 regulated nuclear factor-kappa B (NF-κB) transcriptional activity in CML, we did not observe similar effects in FLT3+ AML cells. Rather, proteomics analyses suggested a role for PSMD3 in neutrophil degranulation and energy metabolism. Finally, we identified additional PSMD subunits that are upregulated in AML patients with mutated versus wild-type FLT3, which correlated with worse outcomes. These findings suggest that different components of the 19S regulatory complex of the 26S proteasome can have indications for OS and may serve as prognostic biomarkers in AML and other types of cancers.
Keywords: FMS-like tyrosine kinase 3 (FLT3); acute myeloid leukemia (AML); proteasome 26S subunit non-ATPase 3 (PSMD3); proteasome inhibition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. doi: 10.1182/blood.V95.11.3489. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

