Involvement of Small Non-Coding RNA and Cell Antigens in Pathogenesis of Extramedullary Multiple Myeloma
- PMID: 36499093
- PMCID: PMC9741227
- DOI: 10.3390/ijms232314765
Involvement of Small Non-Coding RNA and Cell Antigens in Pathogenesis of Extramedullary Multiple Myeloma
Abstract
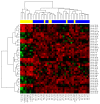
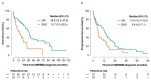
Extramedullary multiple myeloma (EMD) is an aggressive disease; malignant plasma cells lose their dependence in the bone marrow microenvironment and migrate into tissues. EMD is a negative prognostic factor of survival. Using flow cytometry and next-generation sequencing, we aimed to identify antigens and microRNAs (miRNAs) involved in EMD pathogenesis. Flow cytometry analysis revealed significant differences in the level of clonal plasma cells between MM and EMD patients, while the expression of CD markers was comparable between these two groups. Further, miR-26a-5p and miR-30e-5p were found to be significantly down-regulated in EMD compared to MM. Based on the expression of miR-26a-5p, we were able to distinguish these two groups of patients with high sensitivity and specificity. In addition, the involvement of deregulated miRNAs in cell cycle regulation, ubiquitin-mediated proteolysis and signaling pathways associated with infections or neurological disorders was observed using GO and KEGG pathways enrichment analysis. Subsequently, a correlation between the expression of analyzed miRNAs and the levels of CD molecules was observed. Finally, clinicopathological characteristics as well as CD antigens associated with the prognosis of MM and EMD patients were identified. Altogether, we identified several molecules possibly involved in the transformation of MM into EMD.
Keywords: NGS; bioinformatics; immunophenotyping; microRNA; multiple myeloma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
MicroRNA Profiling of Bone Marrow Plasma Extracellular Vesicles in Multiple Myeloma, Extramedullary Disease, and Plasma Cell Leukemia.Hematol Oncol. 2025 Jan;43(1):e70036. doi: 10.1002/hon.70036. Hematol Oncol. 2025. PMID: 39804194 Free PMC article.
-
[MicroRNA Analysis for Extramedullary Multiple Myeloma Relapse].Klin Onkol. 2018 Spring;31(Supplementum1):148-150. Klin Onkol. 2018. PMID: 29808690 Czech.
-
Proteomic analysis of the bone marrow microenvironment in extramedullary multiple myeloma patients.Neoplasma. 2022 Mar;69(2):412-424. doi: 10.4149/neo_2021_210527N715. Epub 2022 Jan 18. Neoplasma. 2022. PMID: 35037760
-
Genetic Abnormalities in Extramedullary Multiple Myeloma.Int J Mol Sci. 2023 Jul 9;24(14):11259. doi: 10.3390/ijms241411259. Int J Mol Sci. 2023. PMID: 37511018 Free PMC article. Review.
-
Extramedullary multiple myeloma.Leuk Lymphoma. 2013 Jun;54(6):1135-41. doi: 10.3109/10428194.2012.740562. Epub 2012 Dec 31. Leuk Lymphoma. 2013. PMID: 23210572 Review.
Cited by
-
MicroRNA Profiling of Bone Marrow Plasma Extracellular Vesicles in Multiple Myeloma, Extramedullary Disease, and Plasma Cell Leukemia.Hematol Oncol. 2025 Jan;43(1):e70036. doi: 10.1002/hon.70036. Hematol Oncol. 2025. PMID: 39804194 Free PMC article.
-
Genetic, epigenetic, and molecular determinants of multiple myeloma and precursor plasma cell disorders: a pathophysiological overview.Med Oncol. 2025 Jun 3;42(7):234. doi: 10.1007/s12032-025-02807-0. Med Oncol. 2025. PMID: 40459694 Review.
-
Detection of early relapse in multiple myeloma patients.Cell Div. 2025 Jan 29;20(1):4. doi: 10.1186/s13008-025-00143-3. Cell Div. 2025. PMID: 39881385 Free PMC article.
References
-
- Kyle R.A., Child J.A., Anderson K., Barlogie B., Bataille R., Bensinger W., Blade J., Boccadoro M., Dalton W., Dimopoulos M., et al. Criteria for the Classification of Monoclonal Gammopathies, Multiple Myeloma and Related Disorders: A Report of the International Myeloma Working Group. Br. J. Haematol. 2003;121:749–757. - PubMed
-
- Rajkumar S.V., Dimopoulos M.A., Palumbo A., Blade J., Merlini G., Mateos M.-V., Kumar S., Hillengass J., Kastritis E., Richardson P., et al. International Myeloma Working Group Updated Criteria for the Diagnosis of Multiple Myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

