Comparative Proteomic Analysis of Transcriptional and Regulatory Proteins Abundances in S. lividans and S. coelicolor Suggests a Link between Various Stresses and Antibiotic Production
- PMID: 36499130
- PMCID: PMC9739823
- DOI: 10.3390/ijms232314792
Comparative Proteomic Analysis of Transcriptional and Regulatory Proteins Abundances in S. lividans and S. coelicolor Suggests a Link between Various Stresses and Antibiotic Production
Abstract
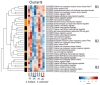
Streptomyces coelicolor and Streptomyces lividans constitute model strains to study the regulation of antibiotics biosynthesis in Streptomyces species since these closely related strains possess the same pathways directing the biosynthesis of various antibiotics but only S. coelicolor produces them. To get a better understanding of the origin of the contrasted abilities of these strains to produce bioactive specialized metabolites, these strains were grown in conditions of phosphate limitation or proficiency and a comparative analysis of their transcriptional/regulatory proteins was carried out. The abundance of the vast majority of the 355 proteins detected greatly differed between these two strains and responded differently to phosphate availability. This study confirmed, consistently with previous studies, that S. coelicolor suffers from nitrogen stress. This stress likely triggers the degradation of the nitrogen-rich peptidoglycan cell wall in order to recycle nitrogen present in its constituents, resulting in cell wall stress. When an altered cell wall is unable to fulfill its osmo-protective function, the bacteria also suffer from osmotic stress. This study thus revealed that these three stresses are intimately linked in S. coelicolor. The aggravation of these stresses leading to an increase of antibiotic biosynthesis, the connection between these stresses, and antibiotic production are discussed.
Keywords: Streptomyces; antibiotics; cell wall stress; label-free protein quantification; nitrogen stress; osmotic stress; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










Similar articles
-
The stringent response is strongly activated in the antibiotic producing strain, Streptomyces coelicolor.Res Microbiol. 2024 May-Jun;175(4):104177. doi: 10.1016/j.resmic.2023.104177. Epub 2023 Dec 28. Res Microbiol. 2024. PMID: 38159786
-
Expression of genes of the Pho regulon is altered in Streptomyces coelicolor.Sci Rep. 2020 May 22;10(1):8492. doi: 10.1038/s41598-020-65087-w. Sci Rep. 2020. PMID: 32444655 Free PMC article.
-
Quantitative Proteomics Analysis Confirmed Oxidative Metabolism Predominates in Streptomyces coelicolor versus Glycolytic Metabolism in Streptomyces lividans.J Proteome Res. 2017 Jul 7;16(7):2597-2613. doi: 10.1021/acs.jproteome.7b00163. Epub 2017 Jun 15. J Proteome Res. 2017. PMID: 28560880
-
The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis.J Antibiot (Tokyo). 2017 May;70(5):534-541. doi: 10.1038/ja.2017.19. Epub 2017 Mar 15. J Antibiot (Tokyo). 2017. PMID: 28293039 Review.
-
Molecular regulation of antibiotic biosynthesis in streptomyces.Microbiol Mol Biol Rev. 2013 Mar;77(1):112-43. doi: 10.1128/MMBR.00054-12. Microbiol Mol Biol Rev. 2013. PMID: 23471619 Free PMC article. Review.
Cited by
-
Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics.Microorganisms. 2023 May 31;11(6):1470. doi: 10.3390/microorganisms11061470. Microorganisms. 2023. PMID: 37374972 Free PMC article.
-
Structural insights into transcription activation of the Streptomyces antibiotic regulatory protein, AfsR.iScience. 2024 Jun 29;27(8):110421. doi: 10.1016/j.isci.2024.110421. eCollection 2024 Aug 16. iScience. 2024. PMID: 39108719 Free PMC article.
-
Expression of Multiple Copies of the Lon Protease Gene Resulted in Increased Antibiotic Production, Osmotic and UV Stress Resistance in Streptomyces coelicolor A3(2).Curr Microbiol. 2024 Dec 17;82(1):43. doi: 10.1007/s00284-024-04021-z. Curr Microbiol. 2024. PMID: 39690306
-
Lydicamycins induce morphological differentiation in actinobacterial interactions.Appl Environ Microbiol. 2025 Jun 18;91(6):e0029525. doi: 10.1128/aem.00295-25. Epub 2025 May 13. Appl Environ Microbiol. 2025. PMID: 40358240 Free PMC article.
-
Metabolic adjustments in response to ATP spilling by the small DX protein in a Streptomyces strain.Front Cell Dev Biol. 2023 Mar 8;11:1129009. doi: 10.3389/fcell.2023.1129009. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36968208 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

